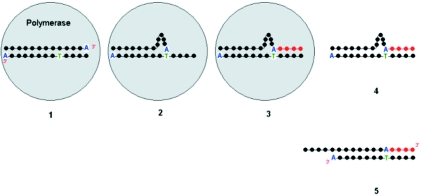
Figure 9.
Model for 5′-CCCCCCCCCCCCA-3′–5′-GGGGTGGGGGGGA-3′ extension. The figure depicts the assumed events during the extension of double-stranded 5′-CCCCCCCCCCCCA-3′–5′-GGGGTGGGGGGGA-3′ oligonucleotide. Polymerase binds the oligonucleotide (1) and shifts A-nucleotide at the 3′ end of 5′-CCCCCCCCCCCCA-3′ until it is paired with T. A single-stranded template-primer fragment and a loop de novo are then formed as a result of the 3′ end slippage (2). The primer strand is synthesized complementary to the template sequence; residues incorporated into the primer are marked in red (3). The enzyme–DNA complex dissociates (4) and a loop relaxes into a structure with overhang at the 5′ end (5). The overhang cannot be used as a template for Klenow exo− due to inability to pair A-nucleotide at the 3′ end of the primer with either nucleotide in sequence of the template.