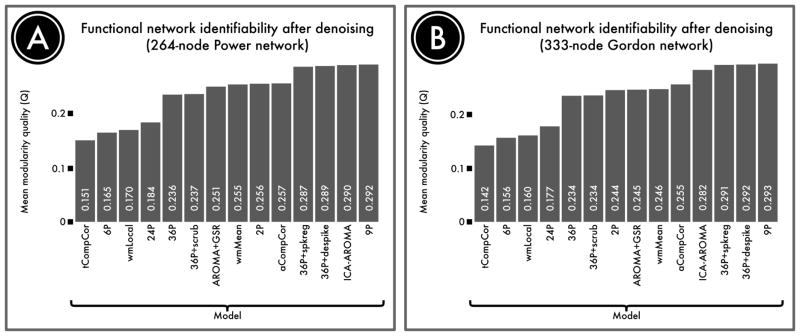
Figure 5. Identifiability of network structure after de-noising.
Although de-noising approaches remove motion artifact from BOLD time series, it is possible that they also remove signal of interest. We quantified the retention of signal of interest as the modularity quality of the de-noised connectome. A, The modularity quality in a 264-node network defined by Power et al. (2011) following confound regression. B, The modularity quality in a second, 333-node network defined by Gordon et al. (2016). ICA-, GSR-, and tissue class-based models performed relatively well, while models that included realignment parameters alone did not remove enough noise to accurately identify network structure.