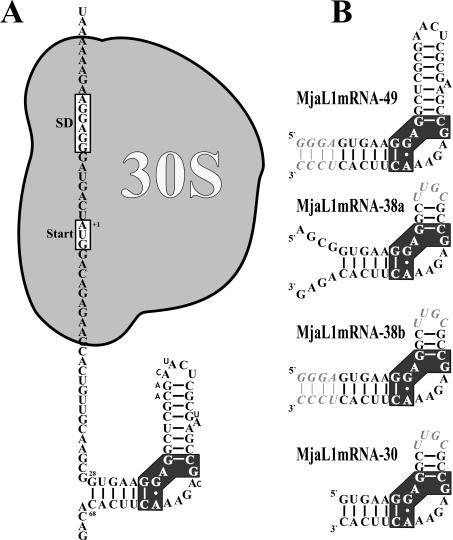
Figure 1.
Secondary structure of the regulatory L1-binding site on the L1mRNA of M.jannaschii and derivatives thereof used in binding experiments and crystallization trials. (A) Localization of the L1-binding site and of the 30S ribosomal subunit as part of the translation initiation complex on the L1mRNA. Nucleotides different in the M.vannielii L1mRNA are shown with smaller sized letters. (B) Fragment MjaL1mRNA-49 comprising nucleotides +28 to +68 with four additional base pairs (italic) at the 3′ and the 5′ end. Fragment MjaL1mRNA-38a comprising nucleotides +24 to +38 and +54 to +72 is closed by the tetraloop UUGC (italic). Fragment MjaL1mRNA-38b is similar to MjaL1mRNA-38a, but the free 3′ and 5′ ends have been replaced by an extended helix as in MjaL1mRNA-49. Fragment MjaL1mRNA-30 comprising nucleotides +28 to +38 and +54 to +68 is closed by a tetraloop as in MjaL1mRNA-38. The cluster of nucleotides conserved in all 23S rRNA and mRNA binding sites is shown with black background.