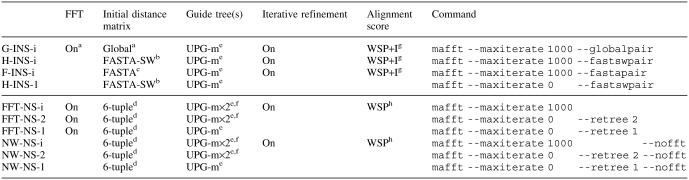
Table 1.
Options of version 5.3 (upper) and the previous version (lower) of MAFFT
aAll pairwise alignments are computed by global alignment with an FFT approximation. The FFT approximation is disabled in the progressive alignment stage.
bAll pairwise alignments are computed with FASTA (25) with the Smith–Waterman optimization.
cAll pairwise alignments are computed with FASTA (25) without the Smith–Waterman optimization.
eUPGMA tree with a modified linkage (for detail see Supplementary Material).
fGuide tree is recalculated based on the first alignment and progressive alignment is re-performed (1,13).
g‘Importance’ (I) value is considered as described in text.
hWSP score is optimized through the iterative refinement (14).