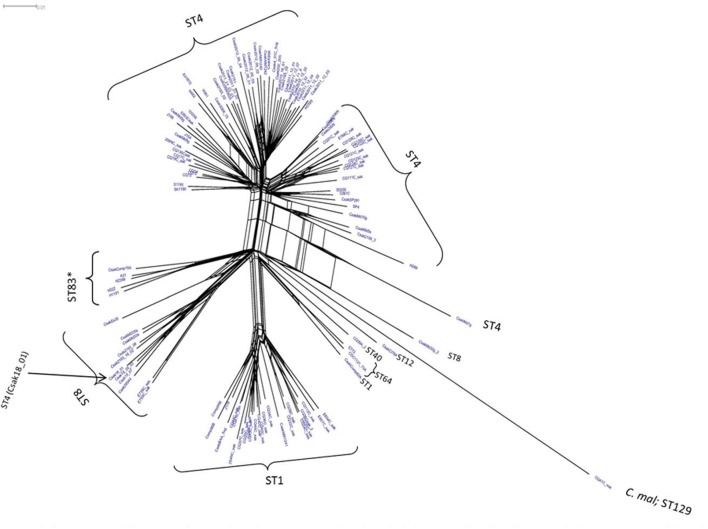
FIGURE 1.
Phylogenetic Neighbor Net (SplitsTree4) analysis of 114 C. sakazakii and phylogenetically related strains, which were generated from the gene-difference matrix (Supplementary Tables S1, S2) and then overlaid with sequence types (ST). The microarray experimental protocol as described by Jackson et al. (2011) and as modified by Tall et al. (2015) were used for the interrogation of the strains for the analysis. The phylogenetic tree illustrates that the Cronobacter microarray could clearly separate the strains according to species and respective ST. The ST83 strains (∗) clustered together along with a single ST83 strain which was obtained from an United States diary powder manufacturing facility. Bar marker represents 0.01 gene differences.