Figure 1.
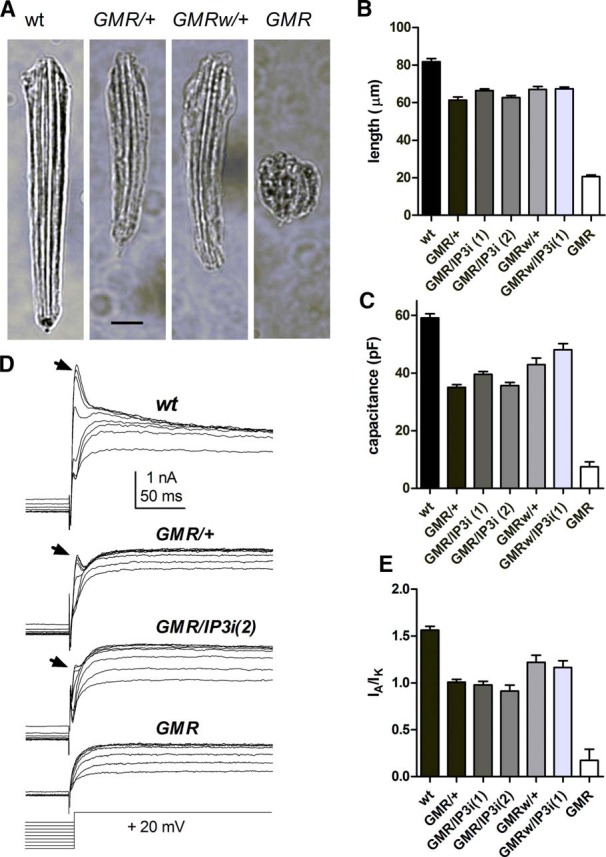
Morphologic and electrophysiological GMRGal4 phenotypes. A, Bright-field micrographs of dissociated ommatidia in wild-type and flies expressing one (GMR/+) or two (GMR) copies of GMRGal4. Scale bar, 10 µm. B, Length of ommatidia in wild type and flies expressing one or two copies of GMRGal4 with or without one or two copies of UAS-IP3R-RNAi (mean ± SEM, n > 10 randomly selected ommatidia from three to four flies per genotype). C, Capacitances in whole-cell recordings from same genotypes (n = 8–34 cells per genotype). Both ommatidia length and capacitance in flies with one copy of GMRGal4 were significantly reduced compared with wild-type (p < 0.001, one-way ANOVA with Dunnett’s multiple comparison test). D, Voltage-sensitive potassium currents at 20 mV after negative 1-s prepulses (–20 to –100 mV) to remove inactivation. Arrows indicate the rapidly inactivating Shaker component (IA), which was greatly reduced in flies expressing one copy of GMRGal4 with or without IP3R-RNAi (two copies) and virtually absent in GMR homozygotes. After 100 ms, the remaining maintained current is largely mediated by delayed rectifier (Shab = IK) channels. E, Ratio of IA (Shaker) peak current to IK (Shab) current measured 100 ms after voltage step: all backgrounds with one copy of GMRGal4 show significantly reduced IA compared with wild-type. Dunnett’s multiple comparison test, p < 0.003, n = 7–17 cells per genotype, except GMRw/IP3R-RNAi (n = 3).
