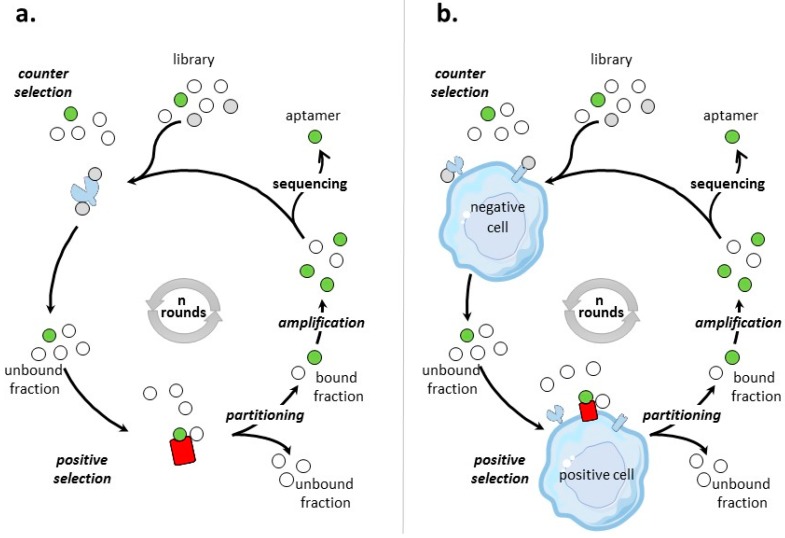
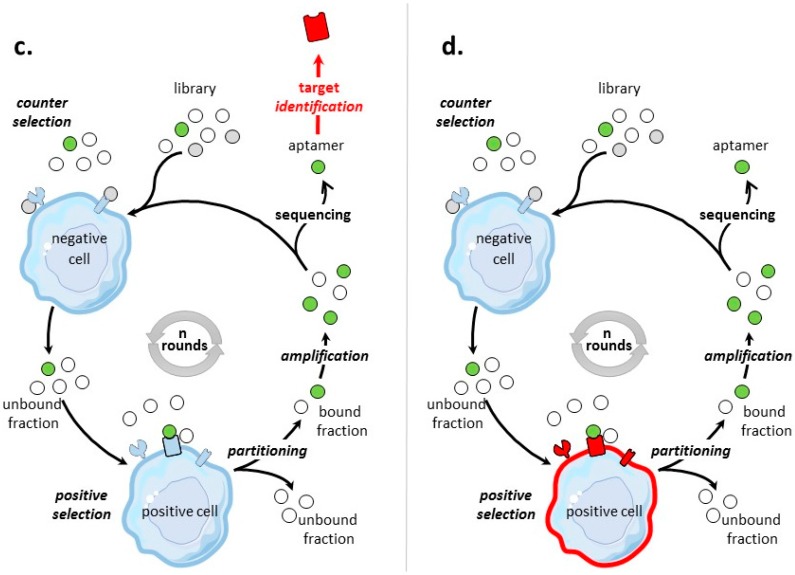
Figure 1.
Scheme of protein- and cell-based SELEX processes. Briefly, the process of SELEX involves first a selection step: the nucleic acid library is incubated with a target (positive selection), which can be preceded or followed by a counter selection phase to remove non-specific nucleic acid molecules. During the partitioning step, bound and unbound fractions are separated. The bound fraction is amplified to obtain an enriched pool for next round of selection. This process is repeated for n rounds until the pool is enriched for sequences that specifically bind the target. These nucleic acid molecules are cloned and sequenced. Individual sequences are aptamers. (a) Protein-based SELEX. The pre-identified purified protein used as target for the protein-based SELEX is colored in red. (b) Cell-based SELEX on a pre-identified tumor cell-surface biomarker colored in red. (c) Cell-based SELEX on a post-identified tumor cell-surface biomarker. The target, identified at the end of the SELEX process, is colored in red. (d) Cell-based SELEX to a tumor cell type. In this case, no particular cell-surface biomarkers are identified and aptamers are specific to the cell’s molecular signature. The whole cell used for positive selection is colored in red.