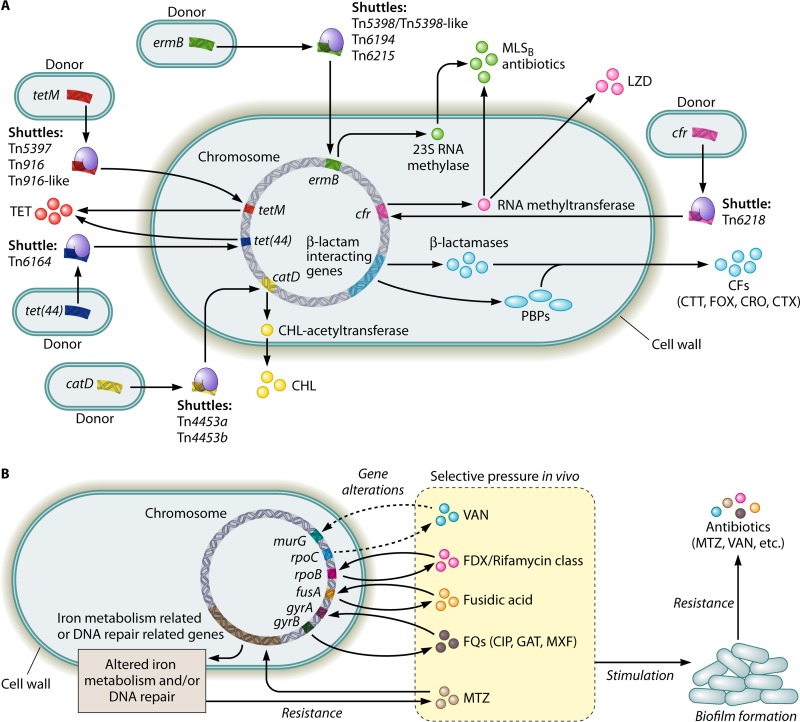
FIG 1.
Diagram illustrating the known factors contributing to the development of antibiotic resistance in Clostridium difficile. (A) Intra- or interspecies transfers of mobile genetic elements via conjugation, transduction, and/or transformation (e.g., transposons) or the natural occurrence of gene mutations (e.g., β-lactamase genes) facilitate C. difficile in obtaining antibiotic resistance genes. (B) Selective pressure in vivo leads to alterations in the antibiotic targets and/or in the metabolic pathways in C. difficile, which on one hand, directly causes antibiotic resistance, while on the other hand, may stimulate biofilm formation. Biofilm formation via different mechanisms (e.g., C. difficile Cwp84, flagella, and the LuxS system) further promotes the development of antibiotic resistance in C. difficile. CFs, cephalosporins; CHL, chloramphenicol; CIP, ciprofloxacin; CRO, ceftriaxone; CTT, cefotetan; CTX, cefotaxime; FOX, cefoxitin; FQs, fluoroquinolones; GAT, gatifloxacin; LZD, linezolid; MLSB, macrolide-lincosamide-streptogramin B; MTZ, metronidazole; MXF, moxifloxacin; PBPs, penicillin-binding proteins; TET, tetracycline; VAN, vancomycin.