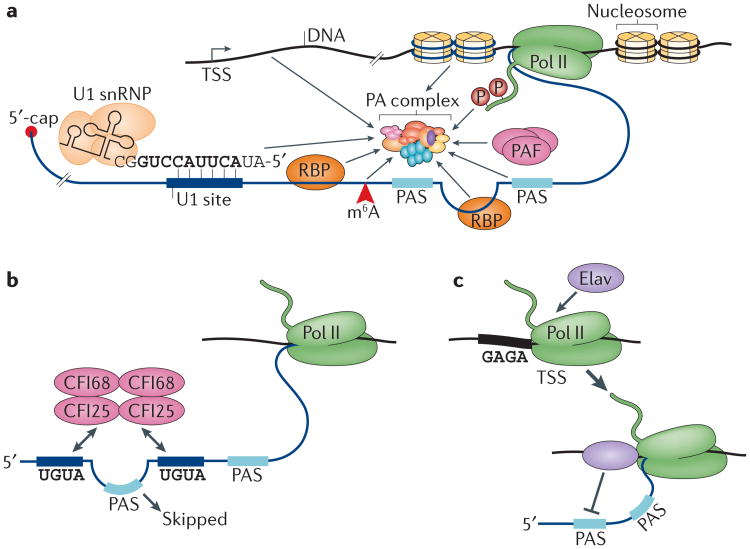
Figure 3. Regulation of APA.
a | The choice of polyadenylation site (PAS) during alternative polyadenylation (APA) can be influenced by various factors, including the gene promoter at the transcription start site (TSS); recruitment of polyadenylation factors directly or of proteins that influence PAS choice; nucleosome density in the region around the PAS; RNA polymerase II (Pol II)-mediated transcription elongation by the Pol II-associated factor (PAF) complex; the function of various RNA-binding proteins (RBPs) associated with the nascent transcript; the presence of N6-methyladenosine (m6A); and inhibition of polyadenylation by the splicing factor U1 small nuclear ribonucleoprotein (U1 snRNP). See the main text for more details. b | A proposed model for the regulation of APA by the cleavage factor I (CFI) complex. Two UGUA elements upstream and downstream of a proximal PAS are recognized by the heterodimeric CFI complex, which consists of CFI 68 kDa subunit (CFI68) and CFI25, leading to skipping of the PAS. c | Regulation of neuronal APA in Drosophila melanogaster by the RBP Embryonic-lethal abnormal visual (Elav). Elav is recruited to Pol II at promoter regions that contain a GAGA sequence, which can cause Pol II pausing. Elav inhibits proximal PAS usage, leading to the expression of long APA isoforms during neurogenesis. PA complex, polyadenylation complex.