Figure 5. Dephosphorylation of SRSF10 by Oxaliplatin.
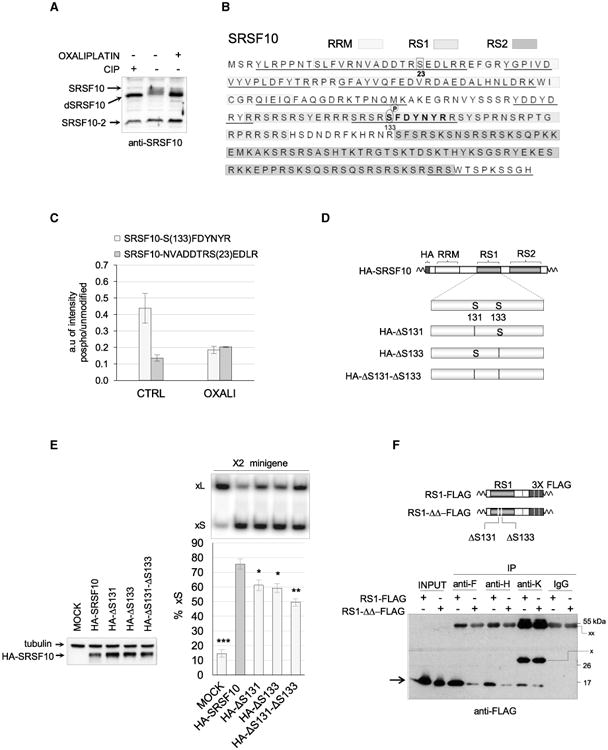
(A) Total cellular extracts were collected 24 hr after treatment or not with oxaliplatin. Aliquots of the untreated cellular extract were incubated with or without calf intestinal phosphatase (CIP) for 15 min at 37°C. Proteins were fractionated on gel and transferred to nitrocellulose to reveal SRSF10. (B) Amino acid sequence of the SRSF10-1 protein showing the different domains (RRM, RS1, and RS2) in differently shaded boxes. The regions of SRSF10 to which peptides identified by LC-MS/MS analysis mapped are underlined.(C) Proteins recovered from duplicate anti-FLAG immunoprecipitations using cells expressing FLAG-SRSF10 and treated or not with oxaliplatin were analyzed by LC-MS/MS analysis after trypsin digestion. Histograms depict the relative abundance of two SRSF10 peptides containing a phosphorylated serine compared to the respective unmodified versions. The peptide in the RS1 domain is shown in bold in (B), and the position of both serines is indicated. (D) Diagram of the mutated versions of HA-SRSF10 carrying either a deletion of S131, S133, or both. (E) The impact of mutated HA-SRSF10 on Bcl-x splicing was tested by co-transfecting minigene X2 and carrying out RT-PCR assays. The percentage of Bcl-xS is represented in histograms with asterisks indicating p values when samples are compared to wild-type HA-SRSF10. Error bars indicate SD. *p < 0.05, **p < 0.01, and ***p < 0.001. (F) The anti-F, anti-H, and anti-K immunoprecipitations used extracts from cells expressing RS1-FLAG or RS1-ΔΔ-FLAG (structure diagrammed on top). The procedure is as described in Figure 3E. A rabbit IgG was used in the control immunoprecipitation.
