Figure 4. Highly transcribed genes influence DNA juxtaposition.
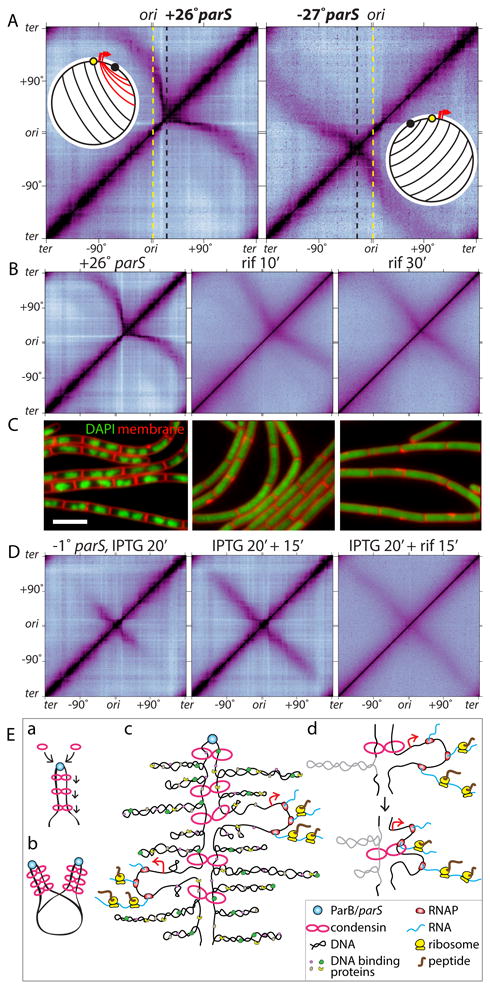
(A) Hi-C contact maps of strains harboring a single parS site at +26° (BWX3403) or -27° (BWX3268). The maps are oriented with the origin at the center of the axes. Dotted lines indicate the positions of the origin (yellow) and the parS sites (black). Schematics show DNA juxtaposition. Red lines highlight the asymmetric juxtaposition adjacent to the highly transcribed genes (red arrows). (B) Hi-C contact maps of the strain harboring the +26° parS site (BWX3403) in the absence or presence of 25 μg/ml rifampicin (rif) for 10 and 30 min. (C) Representative images of cells from (B). DAPI-stained DNA (green) and membranes (red) are shown. Bar, 4 μm. (D) Hi-C contact maps of a strain (BWX3352) harboring a single parS site at -1° and gfp-parB under IPTG control. Cells were induced for 20 min with IPTG, then treated with or without 25 μg/ml rifampicin for 15 min. (E) Schematic model for condensin function. (a) Condensin rings topologically loaded by ParB bound to parS travel down the flanking DNA as handcuffs, (b) resolving newly replicated sister origins by processive loop enlargement. Alternatively, a single ring composed of one or more condensin complexes could encircle the DNA on both sides of parS (not shown). (c) Condensin tethers encounter supercoiled plectonemes, DNA binding proteins, RNA polymerase, and ribosomes translating nascent transcripts. (d) Schematic model of encounters between tethered rings and either a plectoneme (gray) or a transcription complex.
