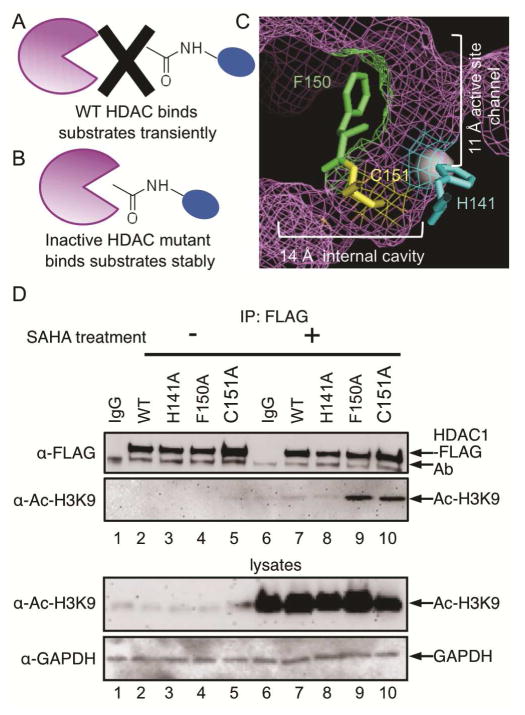
Figure 1.
The Substrate Trapping Strategy. (A) Catalytically active wild type (WT) HDAC enzyme binds substrates transiently. (B) Inactive HDAC mutant enzyme binds substrates more stably, which allows trapped substrates to be isolated by immunoprecipitation. (C) Amino acid residues that were mutated in this study are shown as ball and stick structures in the HDAC1 crystal structure (shown as purple mesh, PBD: 4BKX). The metal ion required for catalysis is shown as a gray sphere. D) FLAG-tagged wild type or mutant HDAC1 were transfected into HEK293 cells, treated with or without SAHA (10 μM) for 24h to induce robust protein acetylation, and then immunoprecipitated with a FLAG antibody, separated by SDS-PAGE, and immunoblotted with FLAG or acetyl-H3K9 antibodies. As controls, lysates prior to immunoprecipitation were separated by SDS-PAGE and immunoblotted with acetyl-H3K9 and GAPDH to assure equal protein content. Additional trials are shown in Figure S1.