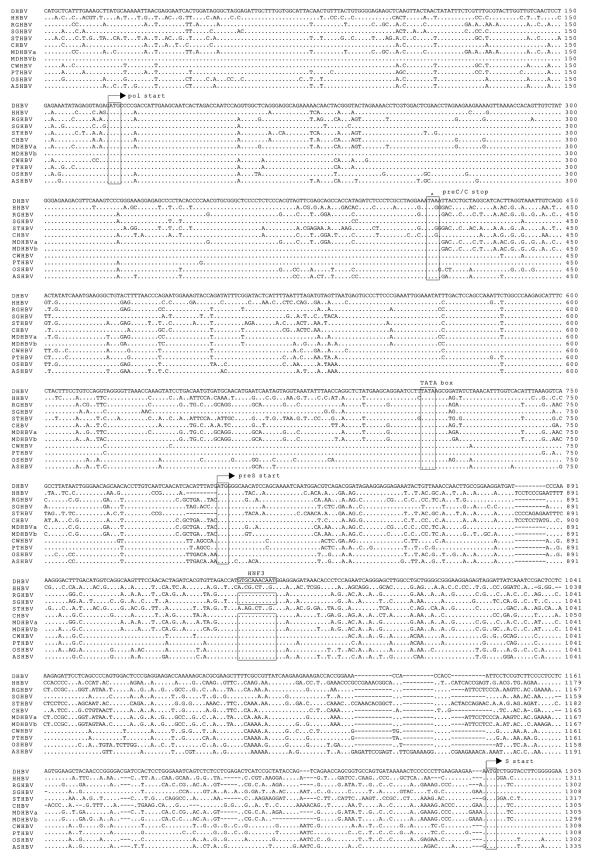
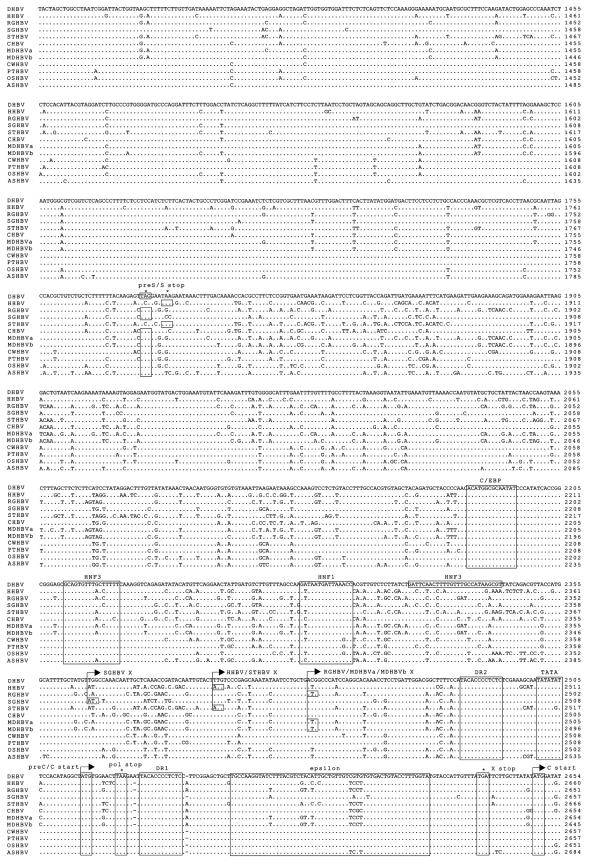
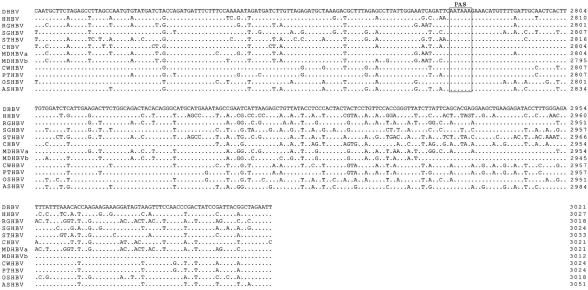
FIG. 1.
Nucleotide sequence alignment with prototypic members of the avihepadnavirus family (DHBV, HHBV, RGHBV, SGHBV, STHBV, and CHBV). Genomes are numbered from an EcoRI sequence in the viral core gene. Dots and dashes represent identical and deleted nucleotides, respectively. Start and stop codons for the viral genes (Pol, polymerase; pre-S/S, envelope proteins; pre-C/C, precore and core; X, X-like protein) are indicated with arrows and asterisks, respectively. Additionally, both features are boxed by rectangles. Selected transcription factor binding sites (HNF, hepatocyte nuclear factor; C/EBP, CCAAT/enhancer-binding protein), the TATA box of the pre-S and core promoter, and other regulatory sequence elements (DR, direct repeats; epsilon; PAS, polyadenylation signal) are also boxed.