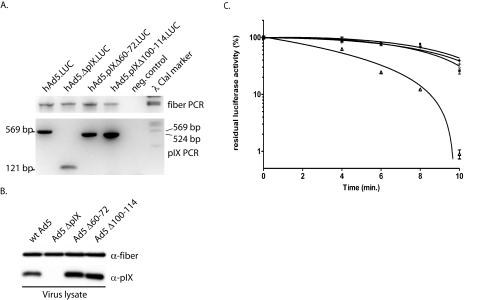
FIG. 3.
Virus stability assay. For the thermostability assay deletions were made in the conserved alanine stretch (Δ60-72) and in the conserved coiled-coil domain (Δ100-114) of the viral backbone. Viruses were propagated on 911 cells and purified by CsCl gradient centrifugation. (A) The presence of the deletions was tested by PCR with primers that flanked the pIX gene in the virus backbone. PCR on hAd5.LUC, which contains wt pIX, should reveal a band of 569 bp, PCR on hAd5.ΔpIX.LUC viruses should reveal a band of 171 bp, PCR on hAd5.pIXΔ60-72.LUC should reveal a band of 528 bp, and PCR on hAd5.pIXΔ100-114.LUC should reveal a band of 525 bp. As internal control, a PCR on the fiber gene was performed with the same samples. Water was used in both PCRs as a negative control. (B) The incorporation levels of the mutant pIX were assayed by Western blotting. As positive and negative controls, wt Ad5 and Ad5 ΔpIX, respectively, were included. (C) Thermostability of wt and mutant viruses. Aliquots of the vectors were incubated in a water bath at 45°C for 4, 6, 8, or 10 min. Residual infectious virus titers were estimated by determining the capacity of the virus to induce luciferase activity in U2OS cell 24 h after infection. The results are presented as percentages of residual luciferase activity. ○, ▵, +, and *, wt hAd5.LUC, hAd5.ΔpIX.LUC, hAd5.pIXΔ60-72.LUC, and hAd5.pIXΔ100-114.LUC, respectively. Each bar represents the cumulative mean ± standard deviation of triplicate analyses.