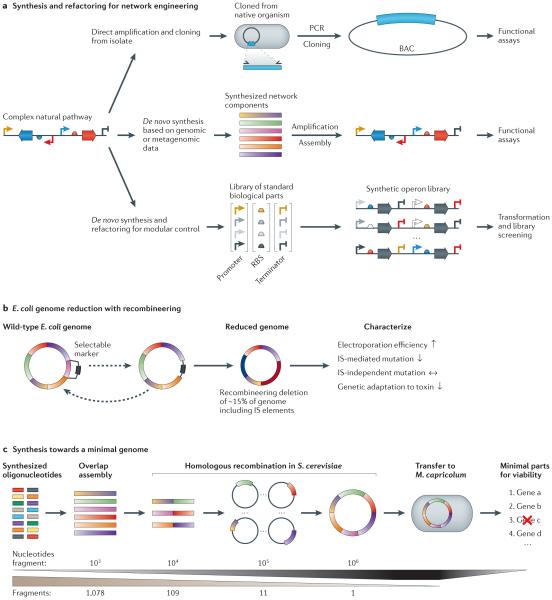
Figure 4. Refactored pathways and minimal genomes.
a | There are numerous approaches to the transfer of gene clusters. Biosynthetic clusters can be cloned from their native organism and transferred to another organism for heterologous expression. For organisms that are recalcitrant to laboratory culture techniques, gene sequences of interest can be identified from genomic or metagenomic databases and subsequently synthesized and introduced into a production organism. The native regulatory elements of a heterologous pathway may not be optimal for expression in the host organism. Refactoring approaches seek to rebuild the pathways using well-characterized modular regulatory elements and removing all native regulation. b | Recombineering (recombination-mediated genetic engineering) was used to delete up to 15% of the wild-type Escherichia coli K12 genome, including insertion sequence (IS) elements. This led to a more stable and streamlined genetic architecture that manifested itself in altered cellular phenotypes. c | The de novo synthesis of the Mycoplasma mycoides genome utilized synthetic DNA oligonucleotides assembled first in vitro and then in Saccharomyces cerevisiae. Following completion of the ~1 Mb genome, the whole construct was transplanted to Mycoplasma capricolum, yielding a viable organism. BAC, bacterial artificial chromosome; RBS, ribosome-binding site.