Figure 3.
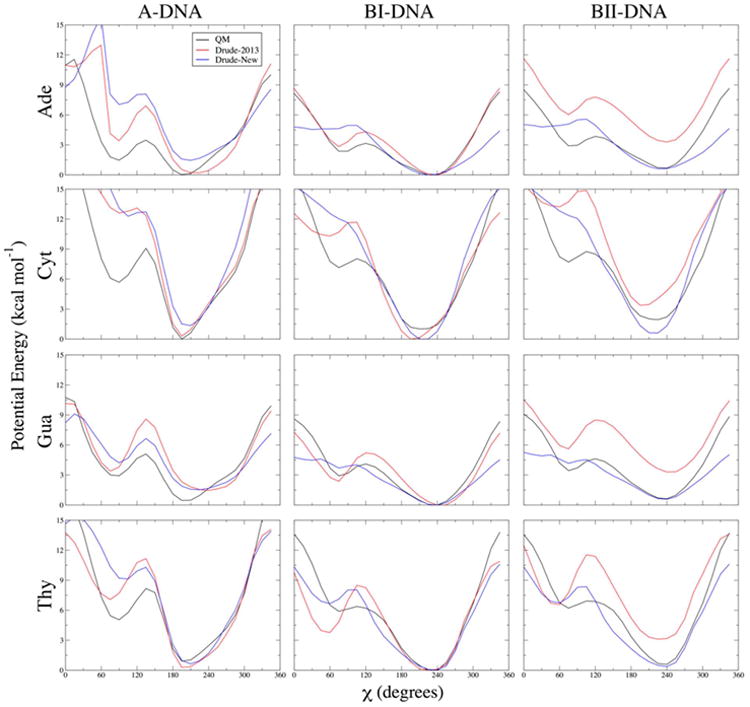
One-dimensional potential energy scans for χ dihedrals in each of the four DNA nucleosides, and each of the three indicated geometries. Backbone β, γ, and ε dihedrals and sugar pucker were constrained to canonical values in each conformational scan. QM potential energies are computed using the RIMP2/cc-pVTZ model chemistry following MP2/6-31G* optimization.
