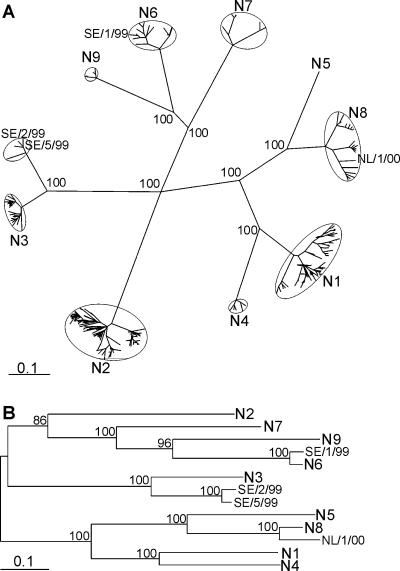
FIG. 5.
Phylogeny of NA genes from Swedish and Dutch black-headed gull influenza A viruses. A tree was generated with all 649 full-length NA amino acid sequences available from public databases (A). Aligned sequences were bootstrapped 100 times, and the Kimura amino acid distance matrices were converted to trees by using a UPGMA clustering algorithm. Very similar trees were obtained when distance matrices were generated by using Hamming distances or the Jones-Taylor-Thornton model (5, 13). The consensus tree was used for recalculation of the branch lengths with the Fitch program of Phylip. Representative nucleotide sequences, when available from Eurasian avian origin, were used to generate a DNA maximum-likelihood tree (B). This tree was built by using 100 bootstraps and 3 jumbles, after which branch lengths were recalculated for the consensus tree. NL/1/00, SE/1/99, SE/2/99, and SE/5/99 represent the NA sequences of black-headed gull viruses of subtypes H13N8, H13N6, H16N3, and H16N3 from The Netherlands and Sweden, respectively. Scale bars roughly represent 10% of changes between close relatives. Small numbers in trees represent bootstrap values.