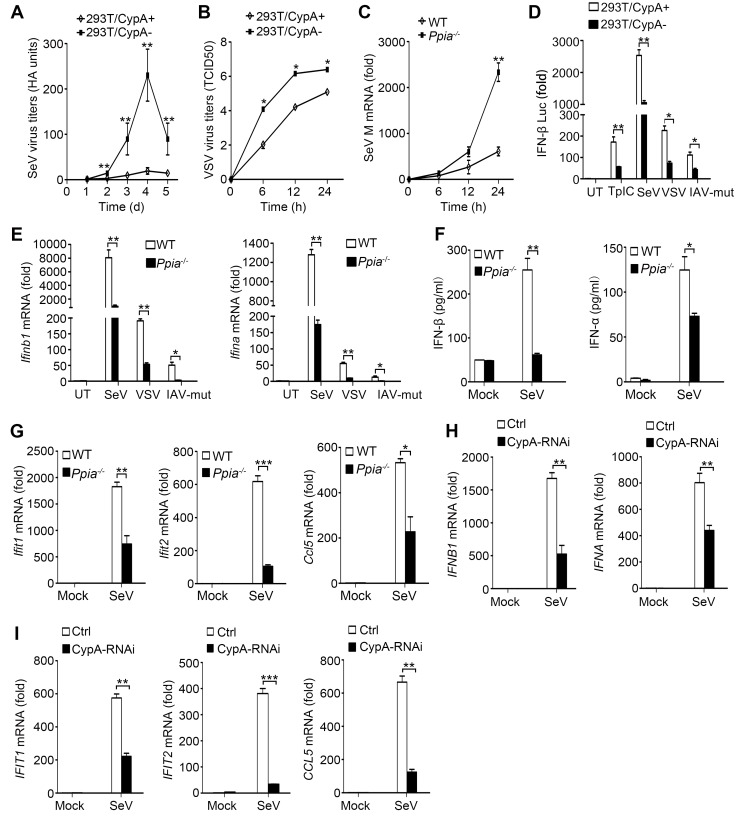
Figure 1. CypA promotes production of type I IFNs against virus infection.
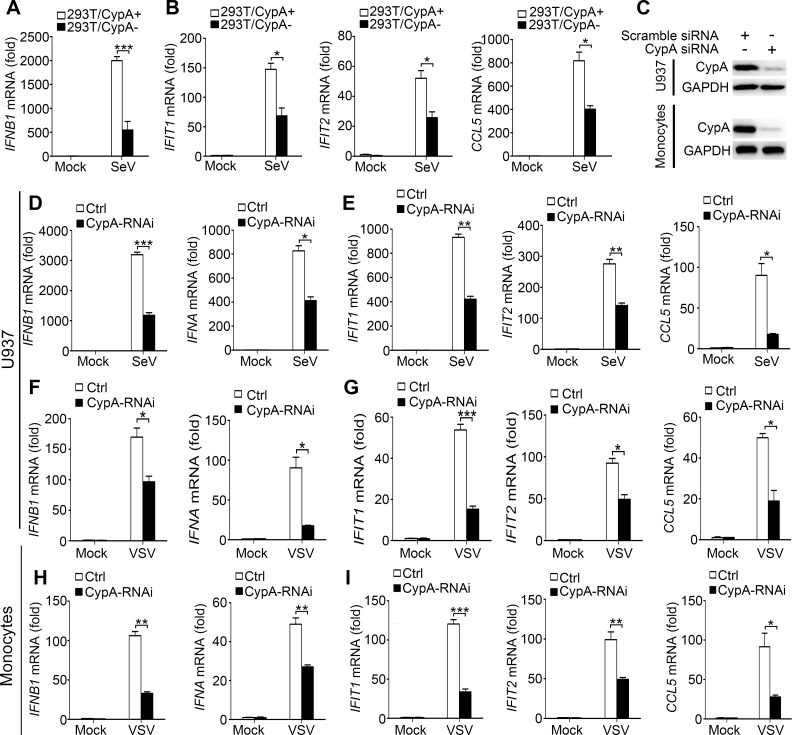
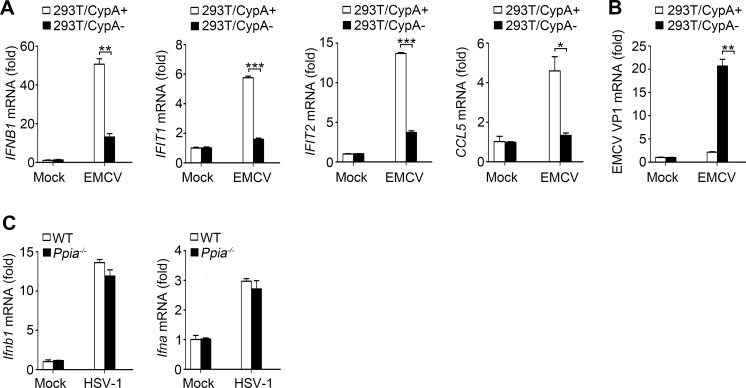
(A) HA assays of SeV in 293T/CypA+ or 293T/CypA- cells infected with SeV (MOI = 1) for the indicated time periods. (B) TCID50 assays of proliferation level of VSV in 293T/CypA+ or 293T/CypA- cells infected with VSV (MOI = 1) for the indicated time periods. (C) Quantitative PCR analysis of SeV M mRNA in wild-type (WT) or CypA-deficient (Ppia−/−) BMDMs infected with SeV for the indicated time periods. (D) Luciferase activity of lysates in 293T/CypA+ or 293T/CypA- cells transfected for 24 hr with IFN-β luciferase reporter (IFN-β-Luc), together with Poly (I:C) (TpIC) or then treated with SeV, VSV, IAV-mut for 6 hr. (E) Quantitative PCR analysis of Ifnb1 and Ifna mRNA in WT or Ppia−/− BMDMs infected with SeV, VSV or IAV-mut for 6 hr. (F) ELISA of IFN-β and IFN-α production in the supernatants of WT or Ppia−/− BMDMs treated with SeV for 12 hr. (G) Quantitative PCR analysis of Ifit1, Ifit2, and Ccl5 mRNA in WT or Ppia−/− BMDMs treated with SeV for 6 hr. (H and I) Quantitative PCR analysis of IFNB1, IFNA (H) IFIT1, IFIT2, or CCL5 (I) mRNA in human monocytes transfected with CypA siRNA or scrambled siRNA for 48 hr and then treated with SeV for 6 hr. Data are shown as mean ± SD (A: n = 5; B-I: n = 3). *p<0.05, **p<0.01, ***p<0.001 (unpaired, two-tailed Student’s t-test). Data are from one representative of at least three independent experiments.
DOI: http://dx.doi.org/10.7554/eLife.24425.003