Figure 4.
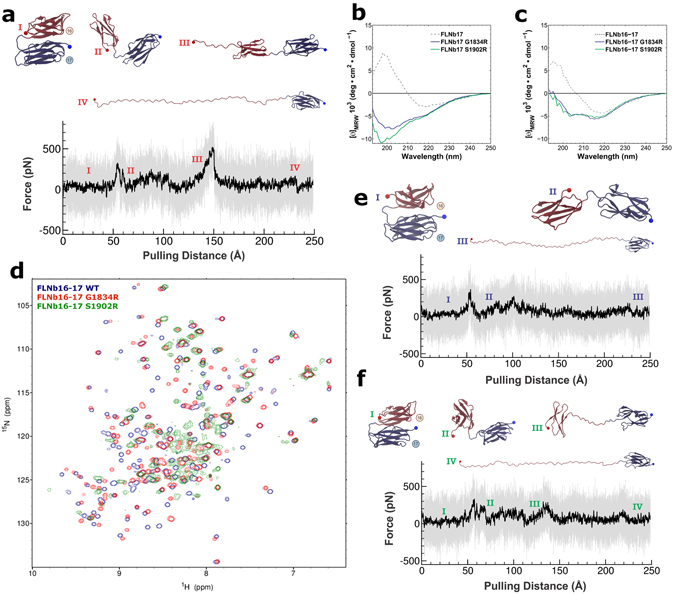
Effects of G1834R and S1902R mutations on FLNb16-17 structure and force response. (a) Unfolding trace of FLNb16-17 WT obtained from SMD a pulling speed of 2.5 Å ns−1 shows that the interaction between domains 16 and 17 is lost first, followed by domain 16 unfolding. The full trajectory is shown gray. The black line represents a moving average with a box size of 500 steps. The snapshots of different time steps are labelled I-IV. (b,c) The CD-spectroscopy analyses of the WT and mutated FLNb17 (b) and FLNb16-17 (c) shows that both G1834R and S1902R destroy the β-sheet folding of FLNb17 and the neighboring domain 16 do not stabilize the β-sheet folding of mutated FLNb17 (c). (d) 1H 15N HSQC spectra of FLNb16-17 WT, G1834R and S1902R show that S1902R has high proportion of unfolded regions, whereas G1834R is mainly structurally ordered having only some disordered regions. (e,f) Unfolding traces of FLNb16-17 G1834R (e) and S1902R (f) obtained from SMD a pulling speed of 2.5 Å ns−1. Similarly as with FLNb16-17 WT (a), the interaction between domains 16 and 17 is lost before domain 16 unfolding. However, with both mutants, forces needed to unfold domain 16 are considerable lower than seen with WT. The snapshots of different time steps are labelled I-IV.
