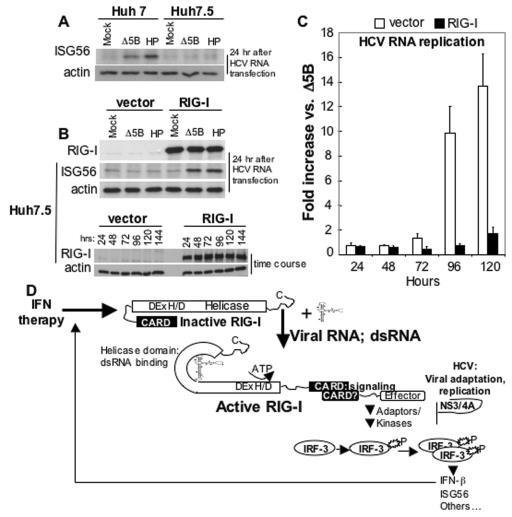
FIG. 5.
Effect of RIG-I on cellular permissiveness for HCV RNA replication. (A) Huh7 and Huh7.5 cells were mock transfected or transfected with Δ5B or HP HCV replicon RNA. After 24 h, the cells were harvested and extracts were subjected to immunoblot for ISG56 and β-actin. (B) Huh7.5 cells were transfected with vector alone or a vector encoding RIG-I. In the upper panel the cells were secondarily transfected, 24 h later, with Δ5B or the HP replicon RNA. After 24 h, cells were harvested, and extracts were subjected to immunoblot analysis for β-actin, ISG56, and the ectopically expressed RIG-I protein. The lower panel shows an immunoblot analysis of β-actin and ectopic RIG-I protein abundance over a 144-h time course posttransfection. (C) Huh7.5 cells were transfected with empty vector (vector) or vector expressing RIG-I. At 24 h, parallel cultures were secondarily transfected with Δ5B or HP HCV replicon RNA containing the firefly luciferase sequence in lieu of Neo, as described in the legend for Fig. 1E. The level of HCV replicon-encoded luciferase activity was measured as a specific marker of HCV RNA abundance (28). The bars show the average fold increase in luciferase expression ± the SD compared to that expressed by the replication-defective Δ5B replicon control. (D) Model of viral RNA binding and IRF3 signaling by RIG-I. Details are given in the text.