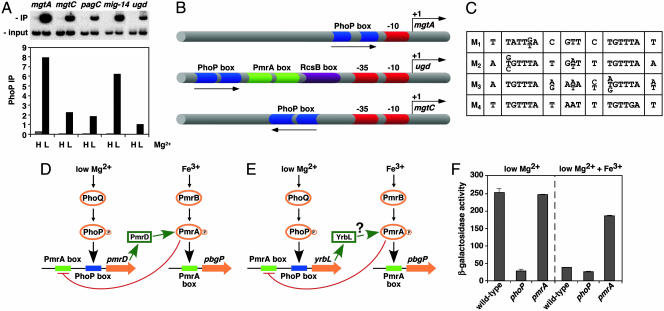
Fig. 1.
Experimental validation of GPS predictions and schematic representation of PhoP-regulated promoters. (A) Chromatin immunoprecipitation (ChIP) of PhoP-regulated promoters predicted by GPS demonstrates binding of the PhoP-HA protein to the PhoP-activated mgtA, mgtC, pagC, mig-14, and ugd promoters, which are found in different profiles. Binding is detected in organisms experiencing low (L) (i.e., 10 μM) but not high (H) (i.e., 10 mM) Mg2+, which correspond to conditions that activate and repress, respectively, the PhoP/PhoQ system. PhoP immunoprecipitation (IP) values correspond to the DNA bound by the PhoP protein (IP) divided by the total DNA before precipitation (input). (B) Representation of PhoP-regulated promoters identified by GPS and experimentally validated. Arrows, transcription start sites; red symbols, RNA polymerase sites; blue symbols, PhoP binding sites; green symbols, PmrA binding sites; purple symbols, RcsB binding sites. Drawing is not to scale. (C) The PhoP binding site submotifs. We built a model for the PhoP binding site by applying an extension of the consensus/patser program (25) by using promoters exhibiting coherent expression patterns (Fig. 3A). This initial model was used for identifying promoter motifs, which were clustered into four subsets based on their degree of sequence similarity to one another (M1–M4). All four submotifs have an hexameric direct repeat separated by 5 bp and preserved those conserved positions critical for PhoP-promoted transcription of the mgtA promoter (13). (D) Model for a multicomponent feedback loop in Salmonella involving the regulatory protein PmrA and its posttranslational activator PmrD. Transcription of the pmrD gene is induced in low Mg2+ in a PhoP-dependent fashion and repressed in the presence of Fe3+ in a PmrA-dependent manner. The PhoP and PmrA proteins exert their regulatory effects directly on the pmrD promoter, which has binding sites for these two proteins. (E) Model for regulatory interactions at the Escherichia coli yrbL promoter, which harbors PhoP- and PmrA-binding sites arranged in a fashion similar to those present in the Salmonella pmrD promoter. (F) Transcription of the yrbL gene is induced in low Mg2+ in a PhoP-dependent fashion and repressed by Fe3+ in a PmrA-dependent manner. β-gal activity (Miller units) of Escherichia coli strains harboring a lac-transcriptional fusion immediately downstream of the yrbL gene was determined in wild type, phoP, and pmrA backgrounds by using organisms grown in N-minimal medium (pH 7.7) and 10 μMMg2+ with or without 100 μMFe3+.