Figure 5.
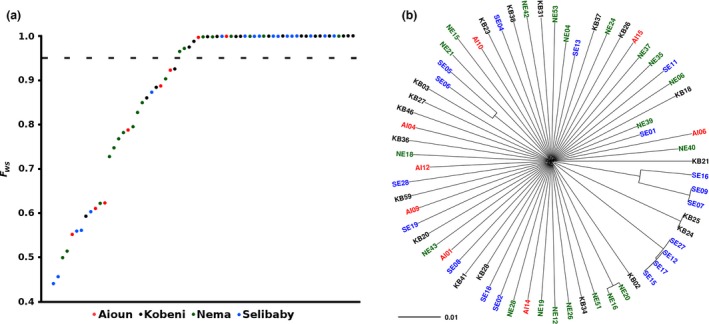
Genomewide sequence analysis of Plasmodium falciparum diversity within and among 65 different infections sampled from Mauritania. The isolates are coloured according to sampling location (red, Aioun; black, Kobeni; blue, Selibaby; green, Nema). (a) Within‐infection fixation indices (FWS) of individual samples show that most are dominated by single genotypes, having FWS values approaching 1.0 (the dashed line indicates the Fws value of 0.95). (b) Neighbour‐Joining tree based on a distance matrix of pairwise SNP identity shows that most of the infections are unrelated and only a minority have similar genotypes. The pairwise distances were calculated using 45 472 biallelic SNPs, and the scale bar indicates the length of a branch corresponding to difference at 1% of SNP positions. Highly related isolates were identified here, and only one of each type was retained for subsequent genomewide analyses to scan for loci with extended haplotypes (isolates excluded were KB24, NE16, SE06, SE07, SE09, SE12, SE15 and SE17). Individual sample information and Accession nos are available on a dedicated project page https://www.malariagen.net/resource/22 and in Table S5 (Supporting information).
