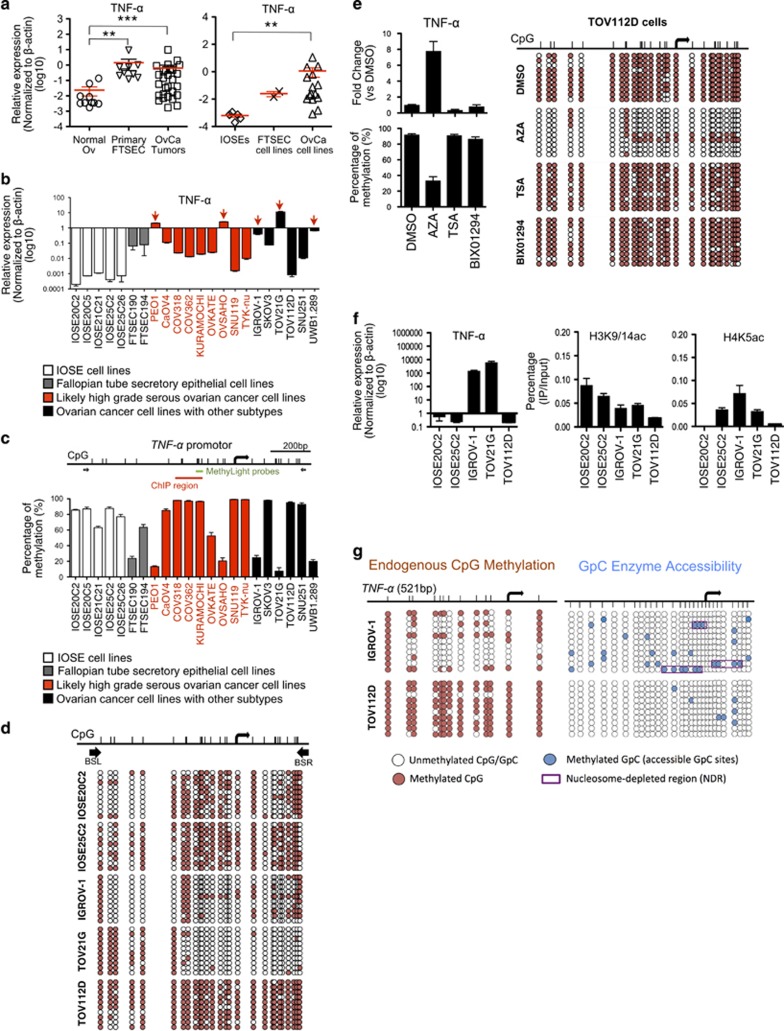
Figure 3.
Promoter DNA hypomethylation and chromatin remodeling regulate TNF-α over-expression in human ovarian cancer cells. (a) The mRNA expression levels of TNF-α was compared between normal ovarian tissue (Normal Ov), micro-dissected fallopian tube secretory epithelial cells from primary normal fallopian tube (Primary FTSEC) and micro-dissected tumor tissue from primary ovarian cancers (OvCa tumors) (left), and between non-malignant hTERT-immortalized human ovarian surface epithelial (IOSE) cell lines, immortalized fallopian tube secretory epithelial cells cell lines (FTSEC cell lines) and human ovarian cancer cell lines (OvCa cell lines)(right), by qRT-PCR. **P<0.01 and ***P<0.001 (Mann-Whitney U test). The data represents means±s.e.m. from three independent experiments. (b) The mRNA expression levels of TNF-α in individual IOSE, FTSEC cell lines and human ovarian cancer cell lines were determined by qRT-PCR. (c) DNA methylation levels of TNF-α promoter in IOSE, FTSEC cell lines and human ovarian cell lines were determined by MethyLight assay using sequence-specific probe (indicated by the green bar). (d) DNA methylation status of TNF-α promoter in IOSE (IOSE20C2 and IOSE25C2) and human ovarian cell lines (IGROV-1, TOV21G, and TOV112D) was investigated by bisulfite genomic sequencing. (White circles, unmethylated CpGs; Red circles, methylated CpGs). Bisulfite-specific PCR primers (BSL and BSR) was indicated by black arrows. (e) The mRNA expression of TNF-α gene and DNA methylation of TNF-α promoter were determined in TOV112D ovarian cancer cells after treatment of 5-aza-2’-deoxycytidine (AZA), trichostatin A (TSA), or methyltransferase inhibitor (BIX01294) for 72 h. DMSO was served as controls. The mRNA expression levels of TNF-α were determined by qRT-PCR (left, upper panel). DNA methylation status was detected by MethyLight assay (left, lower panel) and bisulfite genomic sequencing (right). (f) Chromatin immunoprecipitation (ChIP) was performed in the cell lines with low (IOSE20C2, IOSE25C2, and TOV112D) and high TNF-α expression (IGROV-1 and TOV21G), using antibodies for acetylated H3K9/14 (H3K9/14ac) (middle) and acetylated H4K5 (H4K5ac) (right). Immunoprecipitated DNA was analyzed by qRT-PCR using primers flanking TNF-α promoter region (as indicated by the red bar in (c). Expression of TNF-α transcript in IOSE20C2, IOSE25C2, IGROV-1, TOV21G and TOV112D was shown for reference (left; excerpt from b). (g) Nucleosome-depleted region (NDR) of TNF-α promoter was detected in cell line with high TNF-α expression (IGROV-1), but not in cell line with low TNF-α expression (TOV112D), using high-resolution nucleosome positioning assay (NOMe-Seq). (White circles, unmethylated CpGs/GPCs; red circles, methylated CpCs; blue circles, methylated GpGs; purple boxes, NDR).