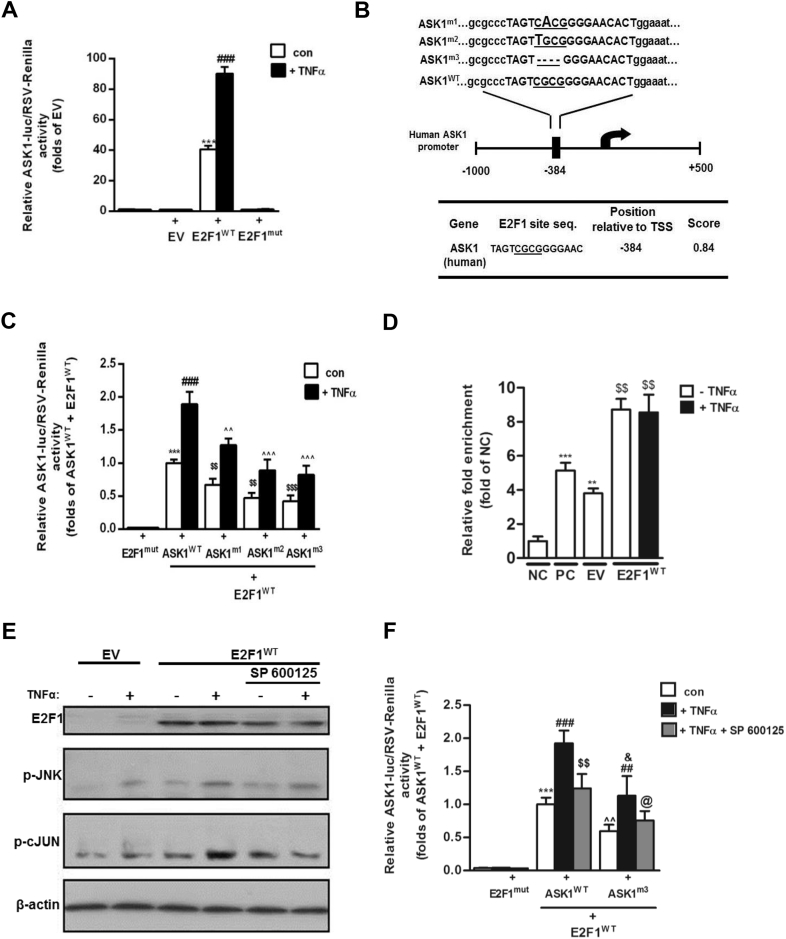
Figure 4.
ASK1 promoter activity is elevated by E2F1 over-expression and JNK phosphorylation input in HEK293 cells. (A) HEK293 cells were transfected with pA3-ASK1-Luc and an empty pcDNA3 (EV) or pcDNA3-E2F1 (E2F1WT) or pcDNA3-E2F1-mut (E2F1mut). After normalization to endogenous control (RSV-Renilla), an empty pcDNA3 (EV) was determined as basal luciferase activity. ASK1-promoter activity was measured after treatment with TNFα (10 ng/ml) for 24 h using Dual Luciferase Assay. Results are mean ± SD of n = 7 independent experiments. ***p < 0.001 con E2F1WT vs. con EV. ###p < 0.001 TNFα 10 ng/ml E2F1WT vs. con E2F1WT. (B) Wild-type (WT) and mutated constructs of predicted E2F1-binding site on ASK1-promoter area by Met-Inspector software. E2F1 binding to the ASK1 promoter is mediated by physical interactions between E2F1 and a core sequence of the binding site (underlined sequence of four nucleotides). Two point mutations (ASK1m1 and ASK1m2) and one deletion mutation (ASK1m3) of this core sequence were made by using mutagenesis kit as elaborated in the Methods, and their effect on ASK1 promoter activity is presented in (C). After normalization to endogenous control (RSV-Renilla), mutated E2F1 (E2F1mut) was determined as luciferase basal activity. ASK1-promoter activity was measured after treatment with TNFα (10 ng/ml) for 24 h using Dual Luciferase Assay. Results are mean ± SD of n = 5 independent experiments. ***p < 0.001 con ASK1WT vs. E2F1mut . ###p < 0.001 TNFα ASK1WT vs. con ASK1WT. $$p < 0.01 con ASK1mutants vs. con ASK1WT. ˆˆˆ/ˆˆp < 0.001/0.01 respectively, TNFα ASK1mutants vs. TNFα ASK1WT. (D) HEK-293 cells were transfected with an empty pcDNA3 (EV) or pcDNA3-E2F1 (E2F1WT) and treated with TNFα for 24 h. Next, chromatin was cross-linked, fragmented and subjected to ChIP with α-RNA poll (as positive control – PC), α-ICAM (as negative control - NC), and α-E2F1 as the protein of interest, as indicated in the Methods. End-point PCR was done using purified DNA and specific primers for E2F1 binding site on the ASK1 promoter; inputs were diluted 1/300 and used as normalization control. Results are mean ± SD of n = 3 independent experiments. **/***p < 0.005/0.01 respectively, vs. NC. $$p < 0.005 vs. EV. HEK-293 cells were transfected with an empty pcDNA3 (EV) or pcDNA3-E2F1 (E2F1WT) and treated with TNFα (10 ng/ml) and SP 600125 (50 μM). Shown are representative blots of Western blot analysis (E) of cell lysates prepared from HEK-293 cells. β-actin was used as a loading control. (F) HEK-293 cells were transfected with pA3-ASK1WT or ASK1m3 and pcDNA3-E2F1-mut (E2F1mut) or pcDNA3-E2F1 (E2F1WT). After normalization to endogenous control (RSV-Renilla), pcDNA3-E2F1-mut (E2F1mut) was determined as luciferase basal activity. ASK1-promoter activity was measured after treatment with TNFα (10 ng/ml) and SP 600125 (50 μM) for 24 h using Dual Luciferase Assay. Results are mean ± SD of n = 3 independent experiments. ∗∗∗p < 0.001 con ASK1WT (E2F1WT) vs. E2F1mut. ###p < 0.001 TNFα ASK1WT vs. con ASK1WT. $$p < 0.005 TNFα + SP ASK1WT vs. TNFα ASK1WT. ˆˆp < 0.005 con ASK1m3 (E2F1WT) vs. E2F1mut. ##p < 0.01 TNFα ASK1m3 vs. con ASK1m3. &p < 0.05 TNFα ASK1m3 vs. TNFα ASK1WT. @p < 0.05 TNFα + SP ASK1m3 vs. TNFα ASK1m3.