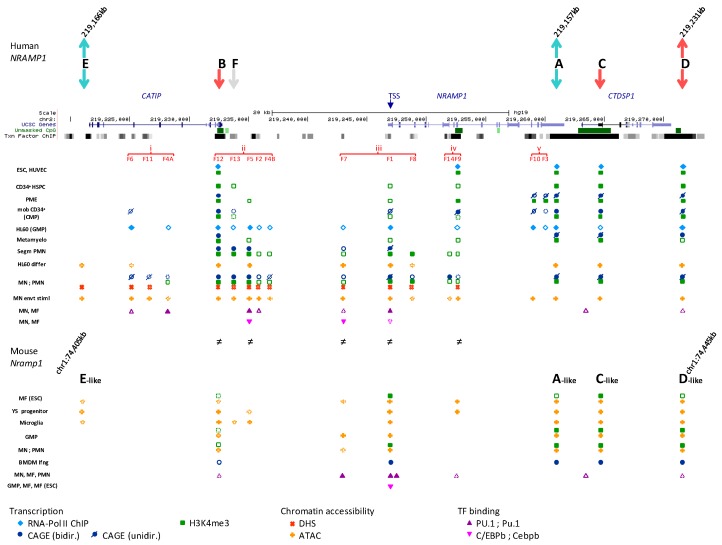
Figure 8.
Recap of transcriptional activity, chromatin accessibility and TF binding at human and mouse Nramp1 loci compiled for selected cell types. Human locus, from top to bottom: CTCF sites (F orientation, red; R orientation, blue; undetermined orientation, grey); coordinates above sites E, A and D indicate potential interaction sites creating regulatory loops; chr2 scale; UCSC genes (names indicated above); CpG islands; ENCODE transcription factor ChIP-seq data. Position of regulatory regions i–v, including DHS F1-F14; cell types: non hematopoietic: ESC and HUVEC; hematopoietic: CD34+ HSPC, mature polymorphonuclear eosinophil (PME), successive stages of myelo-monocytic differentiation, including mobilized CD34+ progenitors (CMP proxy), acute myeloid leukemia HL60 (GMP proxy), neutrophilic metamyelocyte (metamyelo), segmented polymorphonuclear neutrophil (segm PMN), differentiated HL60 cells (PMN, MN, MF), mature MN and PMN, and mature MN stimulated with environmental stimuli (MN envt stiml). ≠ symbols indicate that human and mouse regulatory elements are not functionally equivalent. Mouse locus, from top to bottom: Ctcf sites; hematopoietic cell types: in vitro ESC-derived MF; yolk sac (YS) MF progenitors; microglia; bone marrow GMP, MN and PMN; Ifn-gamma stimulated BMDM. Transcription related signals: CAGE tags, RNA Pol II, H3K4me3, PU.1 and C/EBPb ChIP-seq data and chromatin accessibility data: DNase 1 hypersensitive sites (DHS-seq), Tn5 transposase accessibility (ATAC-seq) are indicated with different symbols and color intensity reflects signal strength.