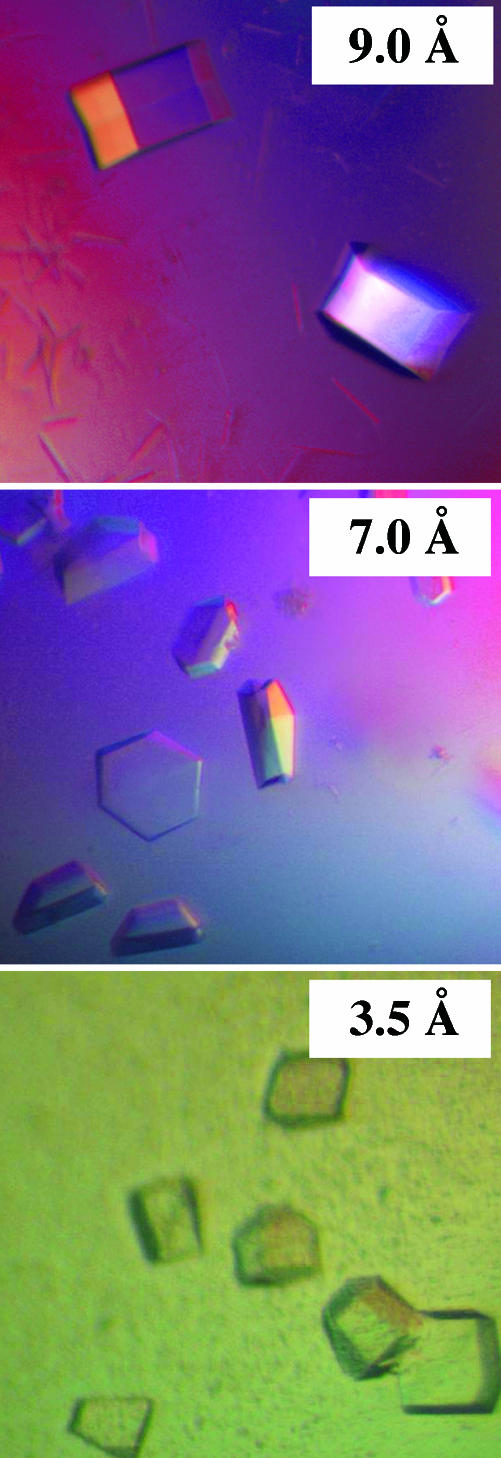
Fig. 3.
Examples of well formed, yet poorly diffracting, crystals obtained with three proteins of NMR grades C or D. The three proteins all were subjected to extensive coarse- and fine-screen crystallization trials (between 17 and 27 fine screens per target), which eventually resulted in the production of macroscopically well formed crystals. However, none of the between 28 and 117 crystals per target examined yielded sufficient resolution for high-resolution structure determination. (Top) A. tumefaciens hypothetical protein Atu1441 (15156516), 17 fine screens, 28 crystals screened; no structure; NMR grade, C. (Middle) S. cerevisiae α-α-trehalose phosphate synthase (YBR126C), 19 fine screens, 100 crystals screened; no structure; NMR grade, D. (Bottom) S. cerevisiae NGG1-interacting factor 3 (YGL221C), 27 fine screens, 117 crystals screened; no structure; NMR grade, C.