Fig. 2.
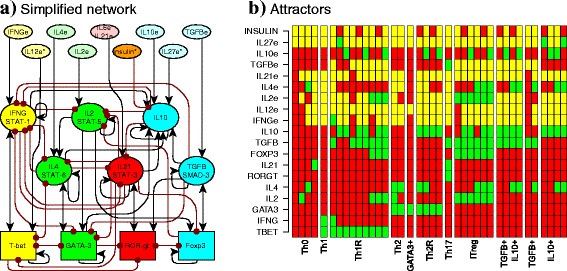
CD4+ T cell regulatory network. a The network includes transcription factors (rectangles), intrinsic cytokines and their signaling pathways (ellipses), and extrinsic cytokines and insulin (ellipses). Node colors correspond to cell types in which each molecule is generally expressed (state = 1): Th1 (yellow), Th2 (green), Th17 (red), iTreg (blue), and insulin (orange). Activations between elements are represented with black arrows, and inhibitions with red dotted arrows. An * is used to indicate the new nodes considered in this network model with respect to that in [13]. b Attractors of the CD4+ T cell regulatory network. Each column corresponds to an attractor. Each node can be active (green) or inactive (red), extrinsic cytokines may be active or inactive (yellow). The following attractors were found in the network: Th0, Th1, Th1R, Th2, GATA3+, Th17, iTreg, TGFβ + IL10+, TGFβ + and IL10+ regulatory cells. Attractors where labeled according to the active transcription factors and intrinsic cytokines
