Fig. 5.
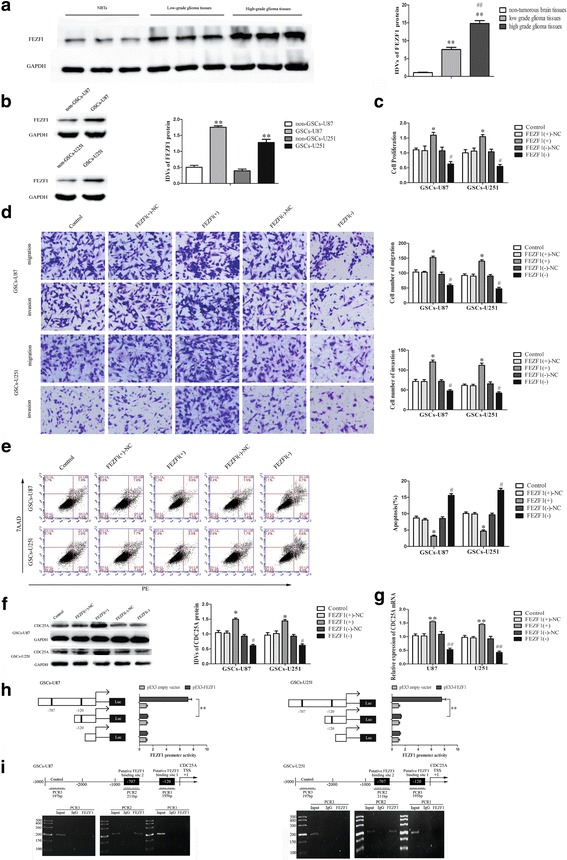
FEZF1 was up-regulated in glioma tissues, and played an oncogenic role in GSCs. a FEZF1 protein expression in non-tumorous brain tissues, low-grade glioma tissues (WHO I-II), and high-grade glioma tissues (WHO III-IV), with GAPDH as an endogenous control. Data are presented as the mean ± SD (n = 10, each group). **P or ## P < 0.01 vs. non-tumorous brain tissue group. b Western blot analysis of FEZF1 expression in non-GSCs and GSCs, with GAPDH as an endogenous control. **P < 0.01 vs. non-GSC group. c CCK8 assay was performed to evaluate the effect of FEZF1 on the proliferation of GSCs. d Quantification of GSC migration and invasion upon FEZF1 over-expression or down-regulation. Representative images and accompanying statistical plots are presented. e Flow cytometry analysis of the effects of FEZF1 on GSCs. Data are presented as the mean ± SD (n = 5, each group). *P < 0.05 vs. FEZF1 (+)-NC group; # P < 0.05 vs. FEZF1(−)-NC group. Scale bars, 20 μm. The photographs were taken at 200 × magnification. f Effect of FEZF1 on the CDC25A protein expression, with GAPDH as an endogenous control. Data are presented as the mean ± SD (n = 5, each group). *P < 0.05 vs. FEZF1(+)-NC group, # P < 0.05 vs. FEZF1(+)-NC group. g Effect of FEZF1 on the CDC25A mRNA expression. Data are presented as the mean ± SD (n = 5, each group). *P < 0.05 vs. FEZF1(+)-NC group, # P < 0.05 vs. FEZF1(+)-NC group. h Schematic depiction of the CDC25A reporter constructs used and the luciferase activity. The Y-bar shows the position of the deletions on the DNA fragments. X-bar shows the constructed plasmid activity after normalization with the co-transfected reference vector (pRL-TK), and relative to the activity of pEX3 empty vector,which the activity was set to 1. Data representmeans ± SD (n = 5, each). i Schematic representation of the CDC25A promoter region 3000 bp upstream of the transcription start site (TSS) which designated as +1. ChIP PCR products for putative binding sites and an upstream region not expected to associate with FEZF1 are depicted with bold lines. Immunoprecipitated DNA was amplified by PCR. Normal rabbit IgG was used as a negative control
