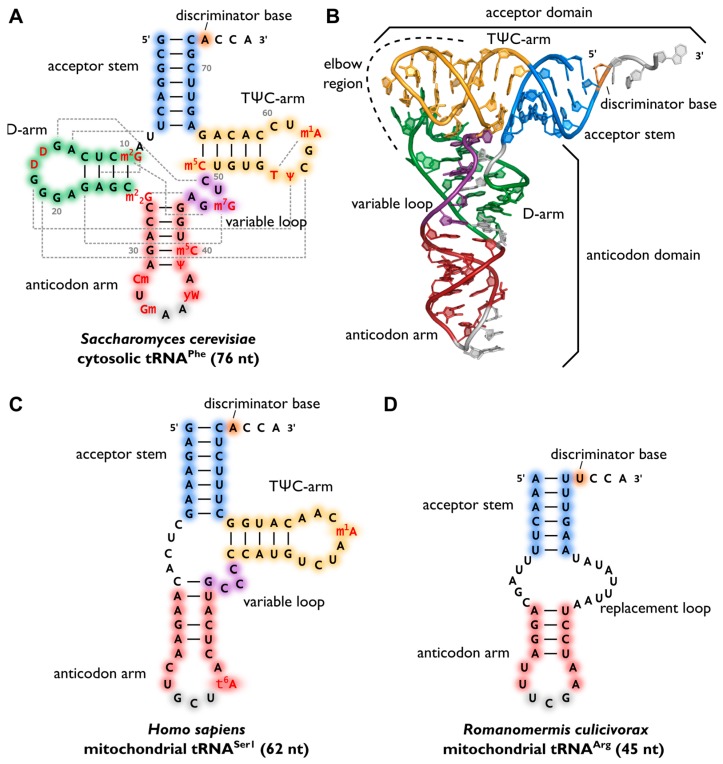
Figure 1.
Variability of transfer RNA (tRNA) structures. (A) The canonical cloverleaf secondary structure of cytosolic tRNAPhe from S. cerevisiae is shown with acceptor stem (blue), D-arm (green), anticodon arm (red), variable loop (purple) and TΨC-arm (yellow). The anticodon is labeled in grey, the discriminator base in orange and post-transcriptional modifications in red. Grey dashed lines indicate tertiary interactions based on structural data [11]. Base numbering corresponds to Sprinzl et al. [25] and length of the RNA is indicated in parenthesis; (B) The L-shaped tertiary structure of the cytosolic tRNAPhe from S. cerevisiae. Protein Data Bank entry (PDB): 1EHZ [11]. The acceptor domain is composed of stacked T-arm and acceptor stem, whereas D- and anticodon arm form the anticodon domain. The region where both domains come together and interact with each other via tertiary base pairing is also called elbow region; (C) Secondary structure of human mitochondrial tRNASer1, which lacks the whole D-arm [26]; (D) Secondary structure of the mitochondrial tRNAArg from the nematode Romanomermis culicivorax, which lacks both D- and T-arm. Instead, we find a so-called replacement loop. It represents the shortest tRNA found in vivo [21].