Figure 1. miR-29 mediates a novel negative feedback loop in SCAP/SREBP-1 signaling and regulates lipid metabolism.
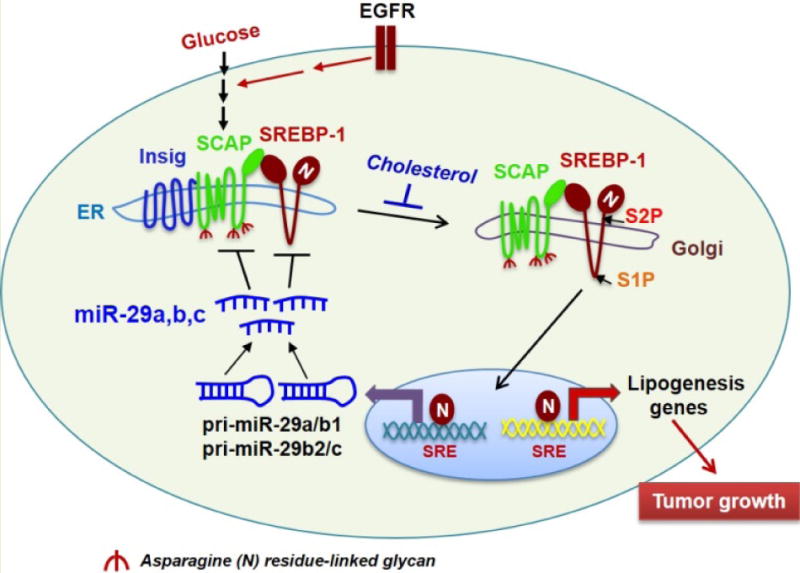
Our previous study showed that glucose-mediated SCAP N-glycosylation enables SCAP/SREBP-1 trafficking from the ER to the Golgi for subsequent proteolytic activation. Furthermore, EGFR signaling enhances glucose uptake, thereby increasing SCAP N-glycosylation and SREBP-1 activation to promote tumor growth [23–25]. High levels of cholesterol increase the association of Insig and SCAP, resulting in the retention of the complex in the ER [6]. Our newly discovered negative feedback loop shows that SREBP-1 transcriptionally activates the expression of pri-miR-29a/b1 and pri-miR-29b2/c, which generate the mature miR-29a, -29b and 29c. In turn, miR-29 reversely inhibits the expression of SCAP and SREBP-1 by binding to their 3′-UTRs, resulting in the downregulation of lipogenesis genes [55]. SRE, sterol regulatory element (SREBP-binding motif present in the promoters of SREBP target genes). S1P, site 1 protease. S2P, site 2 protease.
