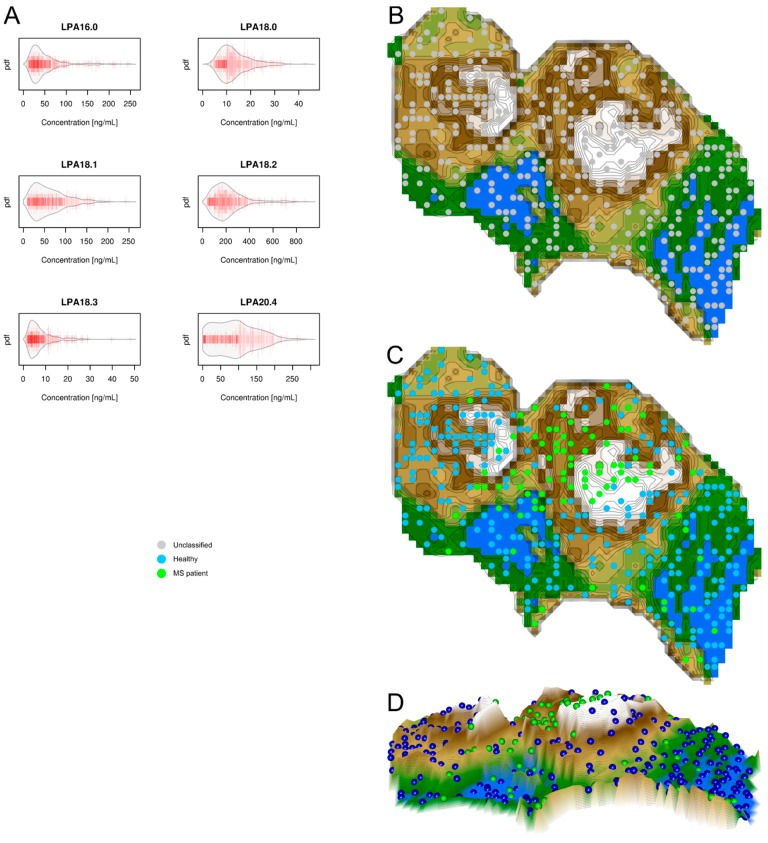
Figure 4.
Data structures of lysophosphatidic acid serum concentrations: Left part, (A) Serum concentrations of d = 6 lysophosphatidic acids (raw data, complete cohort). The data are shown in alphabetical order of lipid mediator names. The beanplots [13] show the individual observations as small lines in a one-dimensional scatter plot, surrounded by a mirrored kernel density estimation of the distributions. Each panel displays a single lysophosphatidic acid marker. LPA = lysophosphatidic acid. Right part: U*-matrix visualization of distance and density based structures of the lysophosphatidic acid serum concentrations (d = 6 lysophosphatidic acid markers) observed in n = 102 multiple sclerosis patients and n = 301 healthy subjects. The figure has been obtained using a projection of the data points onto a toroid grid of 1440 neurons where the opposite edges are connected. The U*-Matrix was colored as a geographical map with brown (up to snow-covered) heights and green valleys with blue lakes. Valleys indicate clusters and watersheds indicate borderlines between different clusters. The dots indicate the so-called “best matching units” (BMUs) of the self-organizing map (SOM), which are those neurons whose weight vectors are most similar to the input. A single neuron can be the BMU for more than one data point or subject, hence, the number of BMUs may not be equal to the number of subjects as in the present case. Differently colored BMUs represent healthy versus MS patient groups; (B) Projection of the markers shown in A onto a self-organizing map. On the raw U-matrix, the BMUs are colored neutrally and no real cluster structure emerges; (C) Analysis of agreement between the marker structure and grouping of the cohort. When the group membership to either the MS patients (green dots) or the healthy subjects (blue dots) is projected onto the U*-matrix, it becomes clear that it does not coincide with the data set’s structure displayed as the U-matrix; (D) A topographic map of the U-matrix visualization of distance and density based structures of the lysophosphatidic serum concentrations. It shows the comparatively weak structure found in the lysophosphatidic acid serum concentrations. No clear ridges-surrounded valleys can be seen; the map mainly consists of “mountains”, which does not allow for concluding a valid cluster structure in the data. Specifically, the beanplots have been drawn using the R package “beanplot” (Kampstra P.; Available online: https://cran.r-project.org/package=beanplot [13]) and the figures displaying geographical map analogies have been created using our R library “Umatrix” (M. Thrun, F. Lerch, Marburg, Germany, Available online: http://www.uni-marburg.de/fb12/arbeitsgruppen/datenbionik//software; file, Available online: http://www.uni-marburg.de/fb12/arbeitsgruppen/datenbionik//umatrix.tar.gz).