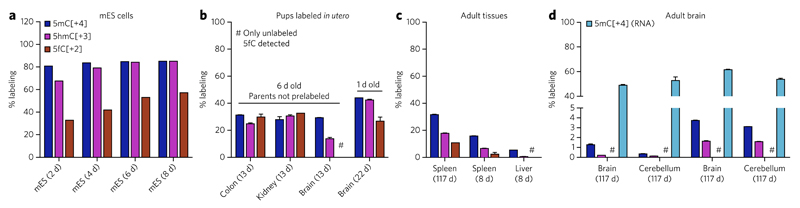
Figure 2. 5fC can be a stable DNA modification in vivo.
(a) Labeling ratios of 5mC, 5hmC and 5fC in the genomic DNA of mEs cells cultured in the presence of [methyl-13CD3]l-methionine. Shown are single measurements; total labeling time is given in brackets. (b–d) Labeling ratios of 5mC, 5hmC and 5fC in the genomic DNA of C57Bl/6 mice fed with the [methyl-13CD3]l-methionine diet. (b) 6-d-old pups labeled from 1 week before birth (total labeling time of 13 d) or 1-d-old newborns (total labeling time of 22 d, parents on labeled diet for 52 d before conception). Shown are mean ± s.e.m. of two animals (6 d old pups) or two technical replicates (1 d old pup). (c,d) Mice labeled in adulthood. Shown are mean ± s.e.m. of at least two technical replicates from individual mice, and total labeling time is shown in brackets. The absence of 5fC[+2] in the brain (d), where 5fC is most abundant (see Fig. 1 and Supplementary Fig. 6), indicates minimal or no further generation of 5fC once placed in postmitotic tissues. Moreover, if 5fC was involved in cycles of methylation and demethylation, its labeling ratio would be similar to that of 5mC in RNA (d). See also Supplementary Figure 9 and Supplementary Table 1.