Figure 7.
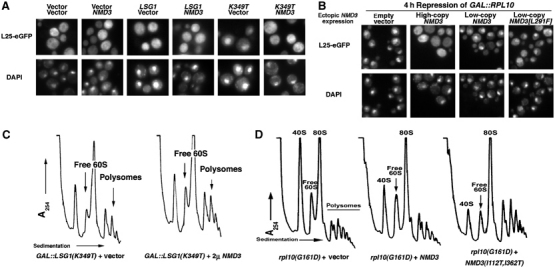
NMD3 suppresses ribosome biogenesis/export defects in lsg1 and rpl10 mutants. (A) Wild-type strain CH1305 was cotransformed with the various combinations of vectors: two empty vectors (pRS425) and (pRS426); pRS425 and pAJ363 (NMD3); pAJ879 (GAL10∷LSG1) and pRS426; pAJ879 and pAJ363; pAJ1109 (GAL10∷LSG1[K349T]) and pRS426; or pAJ1109 and pAJ363. Cells were grown in the presence of galactose for 3 h and prepared for microscopy as described in Materials and methods. (B) DEH221+ (GAL1-10∷RPL10) cells containing pASZ11-RPL25-eGFP (RPL25-eGFP) with either pRS315 (empty vector), pAJ410 (NMD3 2μ), pAJ123 (NMD3 CEN) or pAJ415 (NMD3[L291F] CEN) were grown in galactose-containing medium and diluted four-fold into glucose-containing medium to repress RPL10 expression. After 4 h, cells were prepared for visualization as above. Similar results were obtained with NMD3(I112T, I362T) (data not shown). (C) Cultures of AJY1657 (rpl10[G161D]) cells carrying empty vector (pRS425), pAJ538 (NMD3-myc) or pAJ1315 (NMD3[I112T, I362T]-myc) were shifted to 37°C for 3 h before harvesting. Extracts were prepared in the presence of cycloheximide followed by analysis on sucrose gradients as described in Materials and methods. (D) CH1305 cells carrying pAJ1109 (GAL10∷LSG1[K349T]) with either empty vector (pRS426) or high-copy pAJ363 (NMD3) were cultured in raffinose media followed by induction with galactose. After 4 h, cells were collected, extracts prepared and gradients run as in (C). Arrows indicate increased 60S levels and polysomes in cells coexpressing Lsg1p dominant-negative alleles and high-copy Nmd3p.
