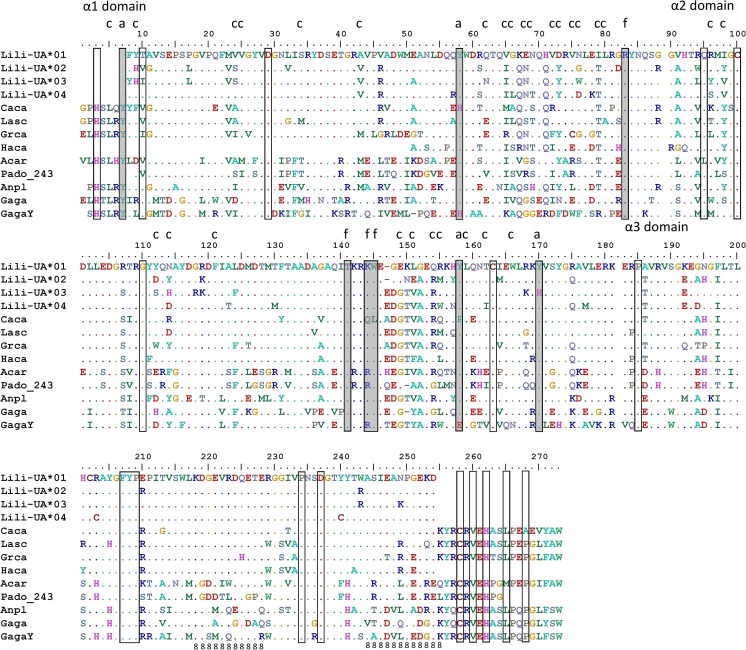
Fig. 2.
Amino acid alignment of Icelandic black-tailed godwit (Limosa limosa islandica; Lili) MHC-I alleles covering the α1, α2, and α3 domains (the first residue of the alignment is the first residue in α1). Identity to Lili-UA*01 is indicated by dots, a blank space determines the end of an exon, and dashes indicate true gaps. Transparent boxes indicate inter- and intra-domain contact residues, light grey boxes indicate peptide main chain sites, c represents non-main chain peptide contacts, a and f indicate the A and F pocket, and 8 indicates the CD8 binding sites (Bjorkman et al. 1987; Saper et al. 1991; Grossberger and Parham 1992; Kaufman et al. 1994; Wallny et al. 2006). NCBI sequence sources are Caca (red knot, Calidris canutus, KC205116), Lasc (red-billed gull, Larus scopulinus, HM025963), Grca (florida sandhill crane, Grus canadensis pratensis, AF033106), Haca (blue petrels, Halobaena caerulea, JF276884), Acar (great reed warblers, Acrocephalus arundinaceus, AJ005503), Pado (house sparrow, Passer domesticus with 3 bp deletion, Pado_243, Karlsson and Westerdahl 2013), Anpl (mallard ducks, Anas platyrhynchos, GU245878), Gaga (chickens, Gallus gallus domesticus BF2, HQ141386), and GagaY (chickens, G. g. domesticus YF6, XM_003643736)