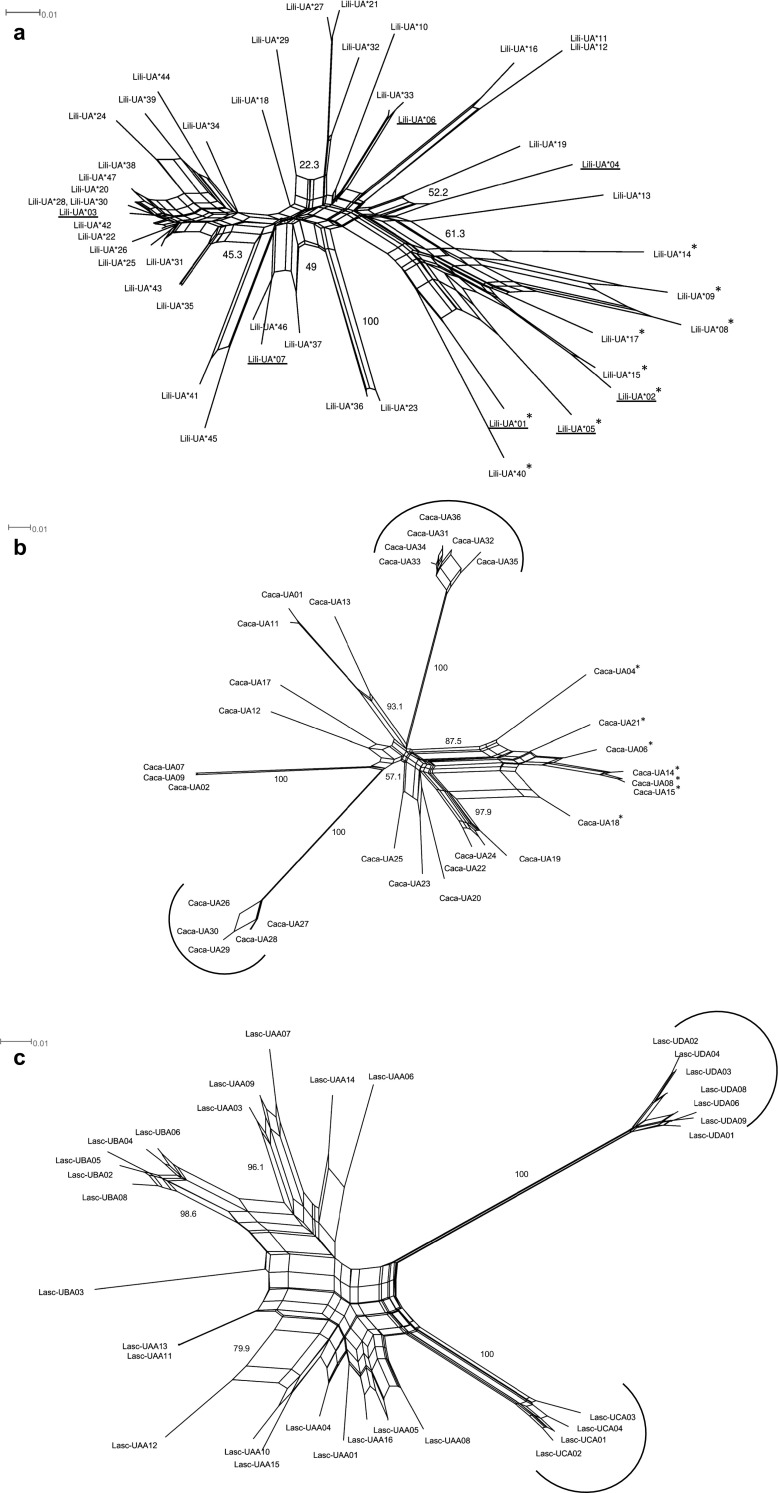
Fig. 4.
Neighbour-net phylogenetic networks of MHC-I exon 3 nucleotide alleles from a Icelandic black-tailed godwits (Limosa limosa islandica), b red knots (Calidris canutus Caca-UA*01-02; Caca-UA*04; Caca-UA*06-09; Caca-UA*11-15; Caca-UA*17-36; Buehler et al. 2013), and c red-billed gulls (Larus scopulinus, Lasc-UAA*01; Lasc-UAA*03-16; Lasc-UBA*02-06; Lasc-UBA*08; Lasc-UCA*01-04; Lasc-UDA*01-04; and Lasc-UDA*06, 08, and 09; Cloutier et al. 2011). The godwit network is based on 47 alleles, and the alleles that are found with both Sanger and Illumina sequencing are underlined. The knot and the gull networks are based on 32 alleles that were downloaded from the GenBank database. Alleles with a 3 bp deletion are indicated by an asterisk, and brackets indicate clades with alleles that have putatively non-classical or pseudogene function. Bootstrap values after 1000 repeats for main splits are presented