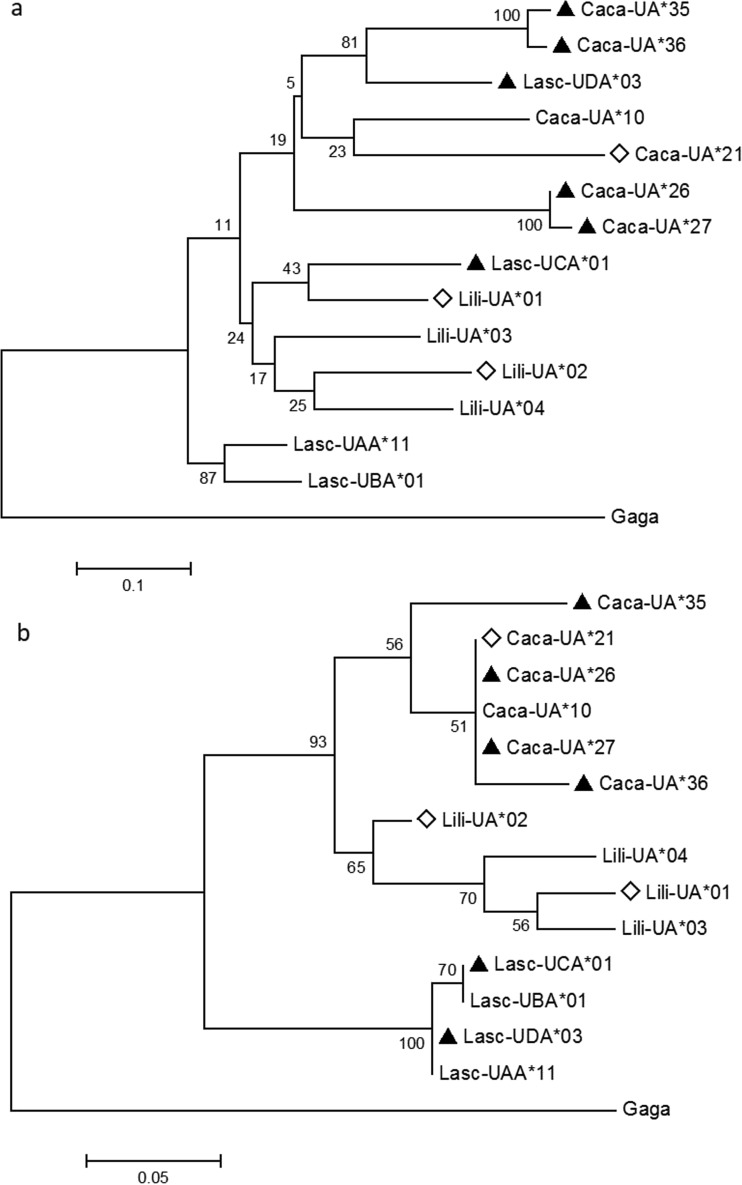
Fig. 5.
Phylogenetic reconstructions using the maximum likelihood method of MHC-I amino acid sequences from three Charadriiformes species, Icelandic black-tailed godwits (Lili, Limosa limosa islandica), red knots (Caca, Calidris canutus), and red-billed gulls (Lasc, Larus scopulinus), and with domestic chicken (Gaga, Gallus gallus domesticus) as outgroup. The trees were built based on a exon 2 and 3 or b exon 4. Putatively classical alleles (Lasc-UAA*11, Lasc-UBA*01, Caca-UA*10, Caca-UA*21, and Lili-UA*01 to Lili-UA*04) without any deletions, are unmarked in the trees, putatively classical alleles with a 3 bp deletion (i.e., short alleles) are indicated with a white diamond (Lili-UA*01, Lili-UA*02, and Caca-UA*21), and non-classical alleles are indicated with a black triangle (Lasc-UCA*01, Lasc-UDA*03, Caca-UA*26, Caca-UA*27, Caca-UA*35, and Caca-UA*36 (Cloutier et al. 2011; Buehler et al. 2013)). Numbers on branches indicate bootstrap values after 1000 repeats