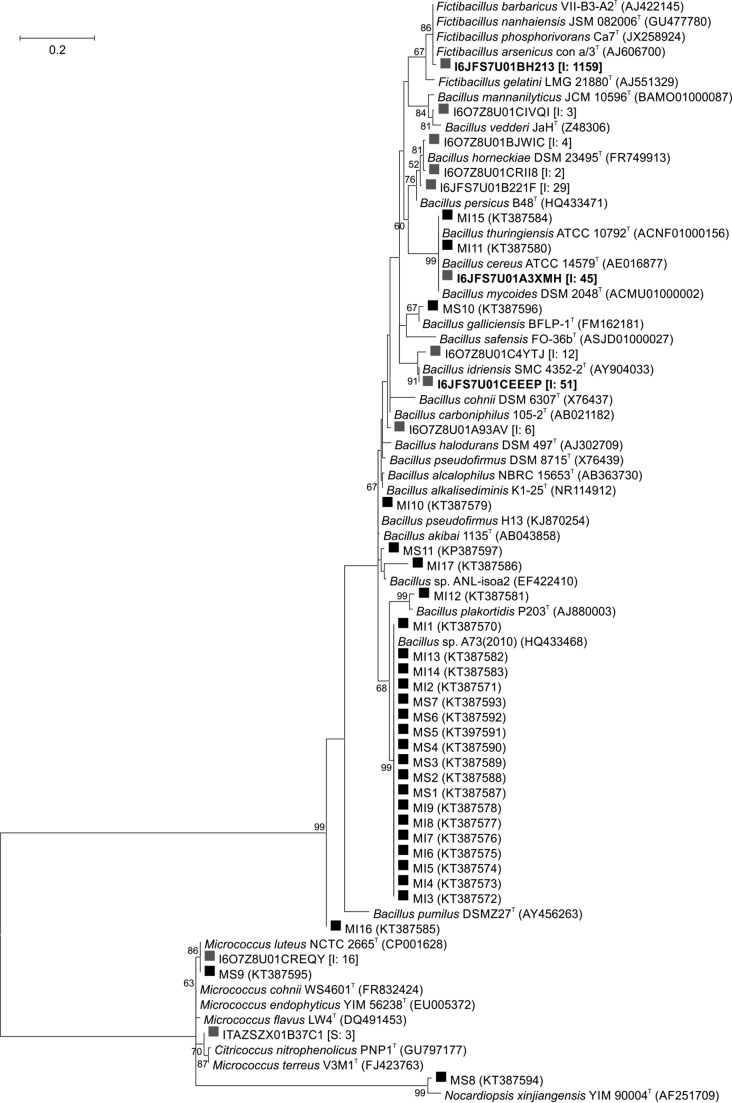
Fig. 6.
Phylogenetic tree of the saline soda lime bacterial isolates and closely related genotypes from pyrosequencing data based on the 16S rRNA gene. Tree was constructed using the Maximum Likelihood method with the Kimura 2-Parameter nucleotide substitution model and is based on 356 nucleotide positions. Type strains are marked with superscript T, GenBank accession numbers are given in parentheses. Bacterial isolates are marked with black squares, while representative sequences obtained by pyrosequencing are marked with grey squares. In the latter case, sequences differing only in one position are represented with a single sequence, number of sequences is given in square brackets (I, interior and S, surface sample) and major phylotypes representing at least 0.5% of total bacterial sequences are highlighted with bold letters