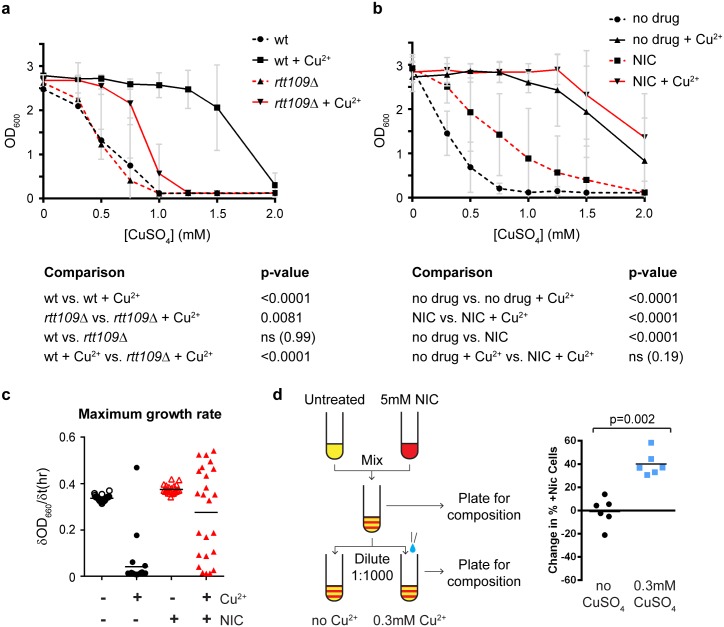
Fig 7. Copper adaptation through stimulated copy number variation (CNV).
a: Copper resistance of 3xCUP1 wild-type (wt) and rtt109Δ cells grown with or without 0.3 mM CuSO4 from Fig 6d. Cells were diluted in media with varying concentrations of CuSO4 and grown for 3 days. Average OD660 is plotted, error bars represent ±1 SD, and n = 6 cultures per condition, each tested at 8 CuSO4 concentrations. p-values were calculated by 1-way ANOVA of area-under-curve values for each culture. b: Copper resistance of 3xCUP1 cells grown with or without 5 mM nicotinamide (NIC) and with or without 0.3 mM CuSO4 from Fig 6c. Analysis as in a; n = 12. c: Maximum growth rate in 0 mM or 0.75 mM CuSO4 of 3xCUP1 cells pretreated with or without 5 mM nicotinamide for 10 generations. dOD660/dt represents the OD change per hour. Four samples each grown with or without nicotinamide were each inoculated in 6 cultures for growth curve determination across 72 hours. Data are the maximum of the first derivative of smoothed OD660 time-course data (see S7c Fig) for each culture. d: Competitive growth assay in 0 or 0.3 mM CuSO4. Two populations of 3xCUP1 cells with different selectable markers were pregrown with or without 5 mM nicotinamide, then mixed and outgrown for 10 generations in direct competition. The graph shows the change in composition of outgrowth cultures across the competition period between inoculation and saturation (10 generations). p-value was calculated by paired t test, n = 6. Raw quantitation data are available in S7 and S8 Data.