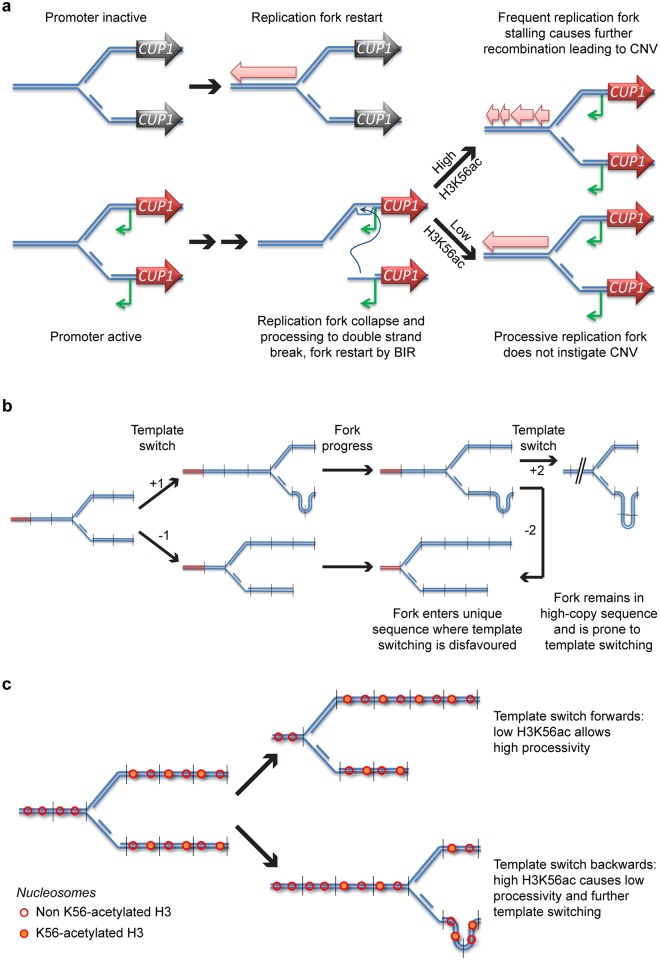
Fig 8. A scheme for stimulated copy number variation (CNV).
a: Proposed mechanism by which promoter activity and acetylated histone H3 lysine 56 (H3K56ac) contribute to stimulated CNV. DNA strands are shown in blue, the inactive CUP1 gene is shown in black, and the induced CUP1 gene is shown in red, with antisense CUT in green. Pink block arrows represent progression of the replication fork. b: Proposed mechanism for CNV asymmetry. DNA strands from repeated DNA are shown in blue, unique sequences are shown in red, and vertical lines indicate repeat boundaries. Numbers indicate the change in copy number for a particular template switch. The result of 2 successive template-switching events with an intervening period of replication are shown, resulting in a 3:1 ratio of contractions to amplifications. c: Additional asymmetry is generated by H3K56ac: nucleosomes around a replication fork (blue) are shown as red circles, either empty or shaded to represent the H3K56ac state. Template switching forward is seen to move the fork to a region of low H3K56ac, whereas template switching backwards moves the fork to a region of high H3K56ac and therefore low fork stability. BIR, break-induced replication.