Fig. 4. IL10VARs display enrichment for differential chromatin in syntenic regions in human tissue and allele specific protein binding.
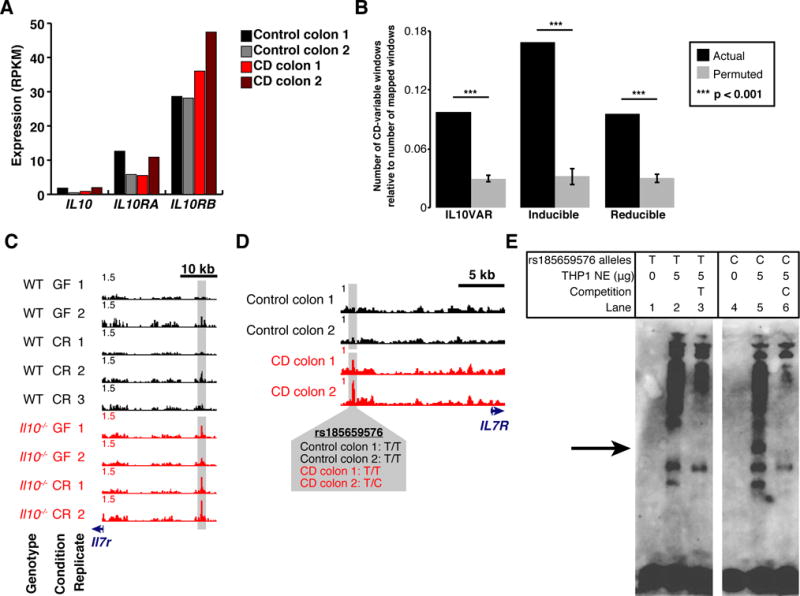
A. Expression levels (RPKM) of IL10, IL10RA, and IL10RB for each human patient biopsied as measured by RNA-seq. B. Regions falling into one of three regulatory element classes from intestinal macrophages were mapped to the human genome and selected based on CD-selective chromatin accessibility in mucosal biopsies from CD and non-CD patients. The number of final CD-variable windows relative to the number of windows that successfully mapped to the human genome is compared to a permuted control for each regulatory element class. P-values were derived empirically based on 1,000 permutations. Error bars represent standard deviation around the mean. C–D. Normalized FAIRE-seq signal at Il7r (mouse, C) and IL7R (human, D) depicting conserved enrichment in mouse intestinal macrophages and human mucosal biopsies. Crohn’s Disease (CD) and control patient genotypes for overlapping variant rs185659576 are also presented. E. Electrophoretic Mobility Shift Assay (EMSA) with human THP-1 macrophage nuclear extract utilizing complementary oligonucleotides containing either the T (reference) or C (alternate) alleles of rs185659576. Protein complex binding specifically to the C allele is demarcated with an arrow.
