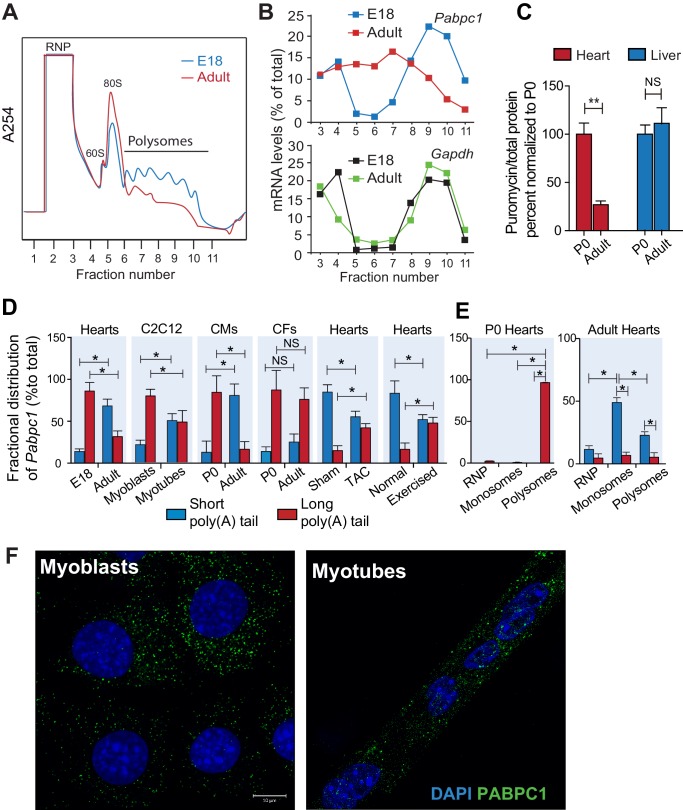
Figure 2. Poly(A) tail length determines cell-type and developmental stage-specific translation of PABPC1.
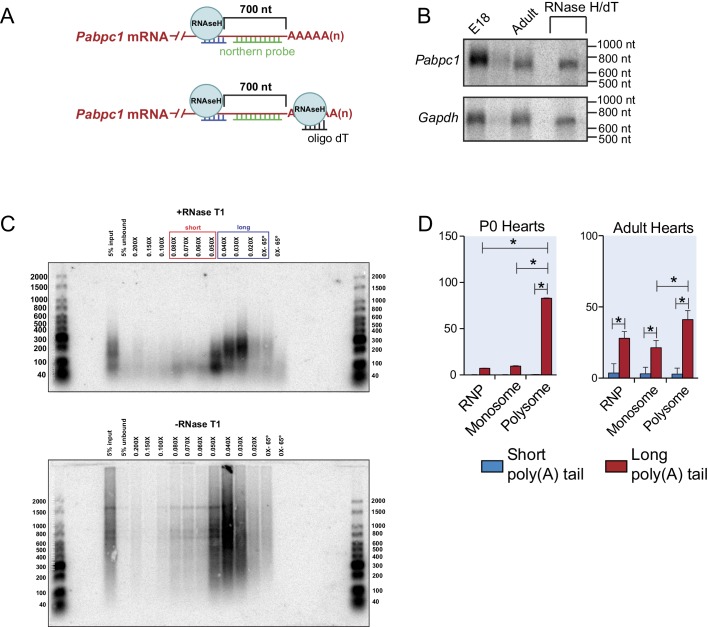
(A). Polysome profile of embryonic day 18 (E18) and adult mouse hearts. (B) Percentage of Pabpc1 and Gapdh mRNAs measured by qPCR in each fraction collected from the polysome profiling. (C) Neonatal and 8-week-old adult wild-type mice were pulsed with puromycin through an intraperitoneal injection. Forty-five minutes following injection, heart and liver tissues were harvested for immunoblotting with anti-puromycin antibody. De novo protein synthesis was quantified as the ratio of puromycin labeled peptides to total protein. (D) Fractional distribution of Pabpc1 mRNAs with short and long poly(A) tails in whole heart, C2C12 cells, cardiomyocytes (CMs), cardiac fibroblasts (CFs), whole heart after TAC surgery, and whole heart after exercise (measured by qPCR following poly(A) tail fractionation). (E) Poly(A) tail length status of Pabpc1 mRNA within P0 and adult heart RNP, monosome, and polysome fractions from sucrose gradients. (F) Pabpc1 single-molecule RNA-FISH in C2C12 myoblasts and myotubes. Data are mean ± s.d (n = 3); *p<0.05, **p<0.005 unpaired two-tailed t-test; NS, not significant.