Figure 14.
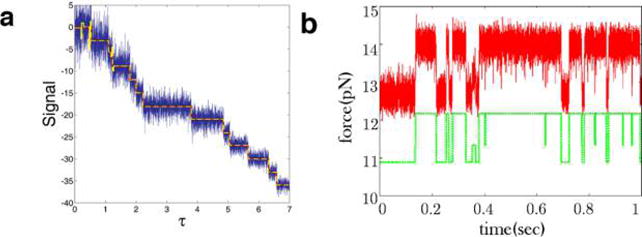
Step-finding algorithms are widely applied across biophysics. (a) In this synthetic time trace containing 15 steps, the fluorescent signal-to- noise ratio decreases with time by an approximate factor of 3. Methods assuming constant noise statistics tend to overfit the start of the trace where the noise level is high (yellow line). A step-finding algorithm that does take into account variable noise performs significantly better (red line which overlaps with the theoretical noiseless trace used to generate this synthetic data).96 (b) An example of a real data trace (red line) showing an RNA hairpin zipping and unzipping (data was obtained through force spectroscopy).268 The offset green line are the steps found using a step-finding algorithm assuming constant noise.269 The time trace in (a) was created via a simple Gillespie algorithm with variable noise ratio. The code implementation of the method in ref 96 can be found online at https://github.com/lavrys/Photobleach. The code for the algorithm that does not take into account variable noise can be found at https://github.com/knyquist/KV_SIC. Adapted with permission from “Single Molecule Conformational Memory Extraction: P5ab RNA Hairpin.” (J. Phys. Chem. B 2014, 118, 6597−6603).268 Copyright 2014, American Chemical Society.
