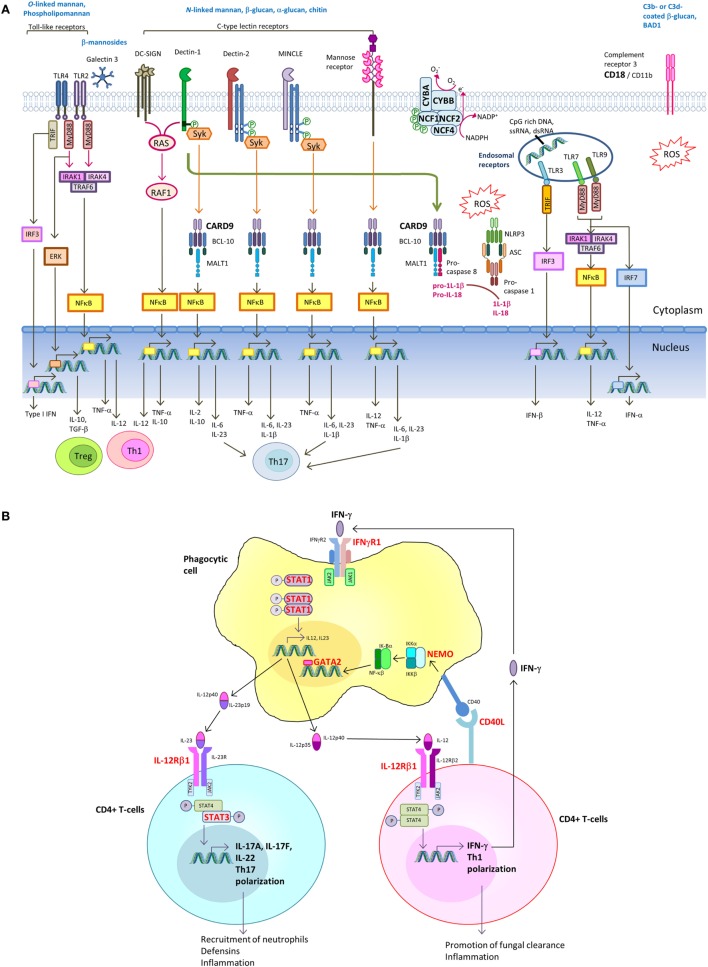
Figure 2.
Signaling pathways in innate recognition of fungal pathogens and differentiation of CD4+ T helper cells. (A) Pathogen-associated molecular patterns (PAMPs) expressed by fungi are recognized by host pattern recognition receptors (PRRs), including toll-like receptors (TLRs), C-type lectin receptors (CLRs) [e.g., dendritic cell (DC)-specific ICAM3-grabbing non-integrin (DC-SIGN), Dectin-1, Dectin-2, MINCLE, and mannose receptor] and complement receptor 3 (CR3). TLRs and CLRs activate multiple intracellular signaling pathways upon binding to specific fungal PAMPs, including β-glucans, chitin, O-linked mannan and N-linked mannan, and nucleic acids. These signals activate canonical or non-canonical nuclear factor-κB and the NOD-, LRR- and pyrin domain-containing 3 (NLRP3) inflammasome. The integration of simultaneously activated PRRs occurs at the level of intracellular adaptors and transcription factors shared between overlapping pathways. The resulting cytokine responses shape the activation of adaptive immune response. Induction of IL-12 drives IFN-γ production by T-helper 1 (Th1) cells, which is crucial for phagocyte activation. Induction of IL-1β, IL-6, and IL-23 promotes Th17 differentiation. Regulatory T-cells (Treg) act as host-driven homeostatic response to keep inflammation under control. (B) Th1 and Th17 differentiation. Polarization of naive T cells into Th1 leads to IFNγ production, and its signaling is mediated through the Janus kinase (JAK)–signal transducer and activator of transcription 1 (STAT1) pathway, leading to transcription of IFNγ-inducible genes. IL-6 and IL-21 upregulate the expression of the retinoic acid-related orphan receptor RORγt and RORα, leading to expression of the inducible component of the IL-23 receptor (IL-23R) and further Th17 development. IL-17A and IL-17F produced by Th17 cells augments neutrophil production in the bone marrow and their recruitment to the site of infection. IL-17A, IL-17F, and IL-22 promote production of antimicrobial peptides in epithelial cells. Molecules in which genetic defects have been identified to be associated with increased susceptibility to endemic mycoses are marked in bold red.