FIGURE 4.
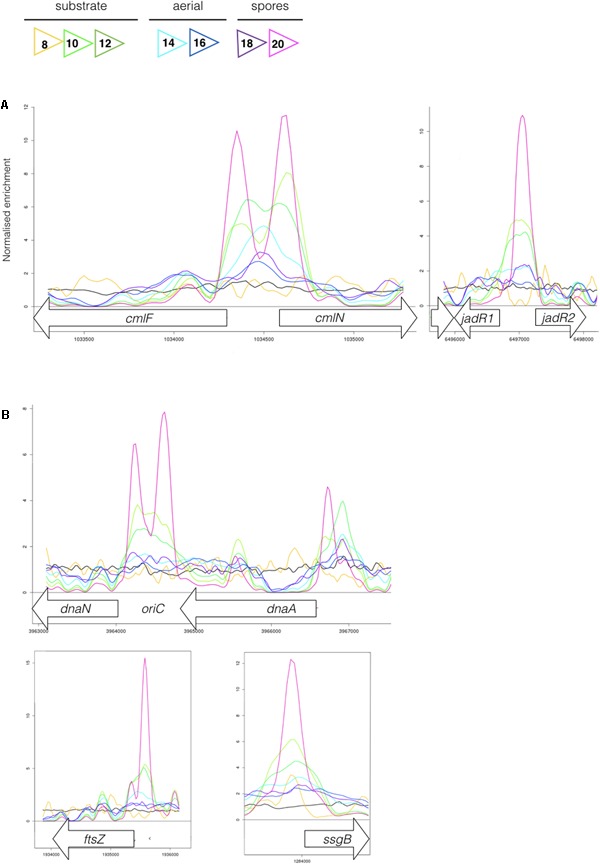
(A) MtrA binds upstream of genes affecting chloramphenicol production. The colored lines show ChIP-seq data for MtrA-3xFlag at 8 h (yellow), 10 h (light green), 12 h (dark green), 14 h (light blue), 16 h (dark blue), 18 h (purple), and 20 h (pink), see Figure 1 for life cycle stages. The black line is the wild-type (negative) control. ChIP peaks are shown at the divergent cmlF and cmlN genes, which encode transporters required for chloramphenicol production (Fernández-Martínez et al., 2014), and jadR1 and jadR2 genes, which encode transcription factors that cross-regulate the jadomycin and chloramphenicol biosynthetic gene clusters (BGCs) (Liu et al., 2013). (B) MtrA binds upstream of genes required for DNA replication and cell division. ChIP-seq peaks, as in Figure 4, upstream of the DNA replication genes dnaN and dnaA and the cell division genes ftsZ and ssgB. In actinomycetes the origin of DNA replication, oriC, is between the dnaN and dnaA genes.
