Figure 4.
The interactions between piRNAs and TEs in the framework of population genetics
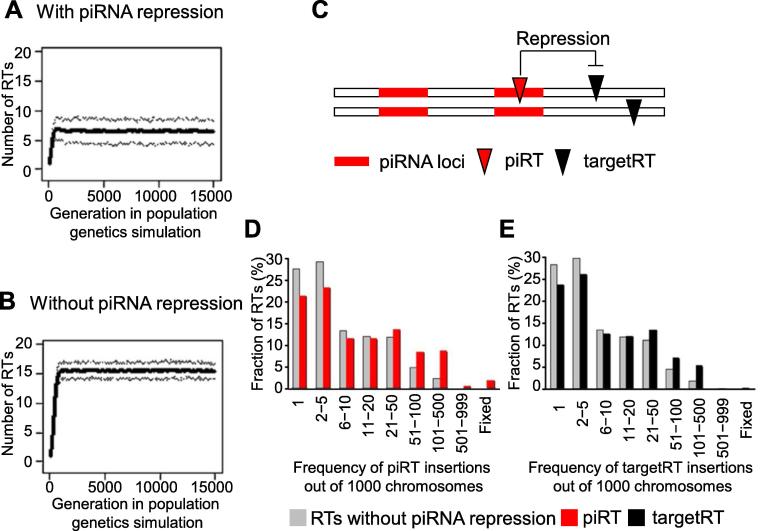
With the same parameter settings in the population genetics simulations, the number of TEs in one chromosome is significantly lower in the presence of piRNA repression (A) compared to the scenario that piRNAs do not repress TEs (B). The solid line represents the mean number of RTs in each chromosome, whereas the thin dashed lines represent the confidence intervals of 90%. C. A schematic illustration of the interaction between piRNAs and TEs. piRTs (RTs in the piRNA clusters, shown in red inverted triangle) refer to the RT that jump into piRNA loci and generate piRNAs to repress RTs of the same family, whereas targetRTs are RTs present outside of piRNA loci, whose activity is reduced by the expressed piRNAs. D. piRTs increase the fitness of the hosts. They are driven by positive selection and spread in the population rapidly, which is manifested by their higher frequencies in the population. E. The frequency of the targetRTs is also skewed toward higher frequencies because their deleterious effects are alleviated by the repression of piRNAs. The figures are adapted from [107]. piRNA, P-element-induced wimpy testis (PIWI)-interacting RNA; RT, retrotransposon; TE, transposable element.