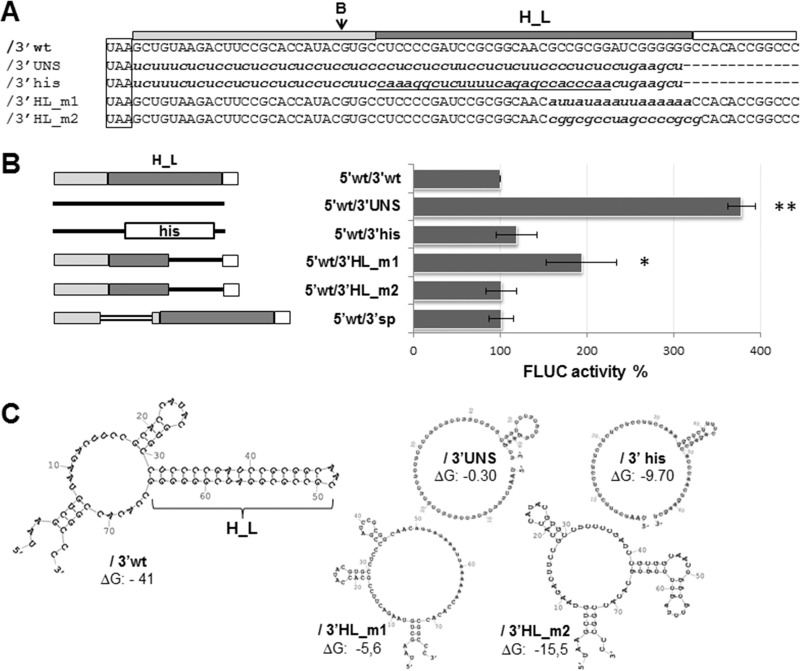
FIG 4.
(A) Alignment of the 5′wt/3′wt 3′ UTR sequence to the corresponding region of mutant mRNAs. The wild-type sequence (/3′wt) is indicated with capital letters, and substitutions are indicated with lowercase letters; the histone 3′-terminal stem-loop sequence is underlined. The position of the BsaAI (B) enzyme recognition site is indicated with an arrow; the H_L sequence is marked with a dark gray box (on top), the stop codon is boxed. (B) (Left) Schematics of the 3′ UTR of wild-type and mutant mRNAs. The 27-nt sequence proximal to the stop codon (light gray box), H_L (dark gray box), and the terminal 12-nt sequence (empty box) are indicated. Substitution sequences are represented with solid lines, except for the histone 3′ terminal stem-loop (his box). The inserted sequence in 5′wt/3′sp is depicted with a double line. (Right) Translation of mutant mRNAs. BSR cells were transfected with the indicated capped mRNAs; FLUC and RLUC activities were determined in cell lysates 8 h later. Mean normalized FLUC values (±SD) from three independent experiments (each conducted in triplicate) are shown as a percentage of those corresponding to 5′wt/3′wt mRNA, taken as 100% (*, P < 0.05; **, P < 0.005). (C) Wild-type and mutant mRNA 3′ UTRs. Predicted secondary structures were obtained by using mfold (46). Free energies (ΔG) are indicated in kilocalories per mole.