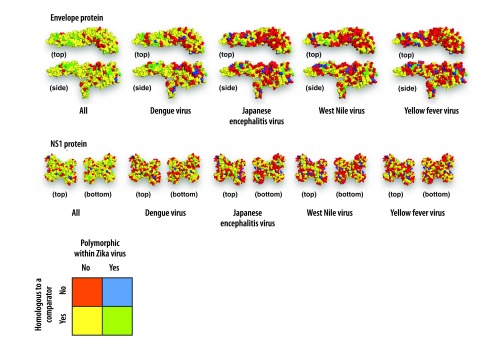
Fig. 3.
Mapping per-site identity and polymorphism onto the structures of Zika virus envelope protein and nonstructural protein 1 dimer
NS1: nonstructural protein 1.
Notes: Sites polymorphic within Zika virus or with high identity between Zika virus and other viruses could compromise respectively sensitivity and specificity of a serological test. Thus the most useful sites are expected to be those that lack identity with other viruses and lack polymorphism within Zika virus, shown in red. Identity between a comparator virus and Zika virus is shown in yellow; polymorphism within Zika virus is shown in blue; and sites with both properties are shown in green. The side view of the envelope protein shows the two transmembrane helices on the bottom. These helices likely remain buried within the lipid bilayer envelope and hence are unavailable for interactions with antibodies. In the structures including all viruses, sites are shown as homologous if they are homologous between Zika virus and any of the flaviviruses. Homologous regions are larger for dengue virus because sites are shown as homologous if they are homologous between Zika virus and any of the four dengue virus serotypes.