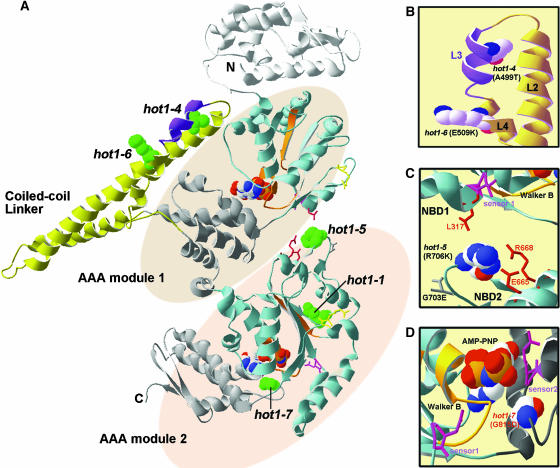
Figure 8.
Location of the hot1 Mutations on the T. thermophilus ClpB Structure.
(A) The available structure of the T. thermophilus ClpB monomer (PDB file 1QVR; chain in [A]) is shown with the coiled-coil domain and AAA modules indicated. Within each AAA module, the NBD domain is colored blue, with the Walker A and B motifs in orange, sensor 1 in pink, and the Arg finger in yellow (see also Supplemental Figure 1 online). The small, α-helical domain of each AAA module is shown in gray. Nucleotide (AMP-PNP) is space-filled in CPK coloring (hydrogen, white; oxygen, red; phosphorous, orange; sodium, blue). Motif 1 as defined by Schirmer et al. (1996) is colored purple in the coiled-coil domain. The residue corresponding to each of the hot1 mutation sites is space-filled and colored green.
(B) Alternative view of the motif II region of the coiled-coil domain indicating the L2, L3, and L4 helices and the residue positions of the hot1-4 and hot1-6 mutations (in CPK coloring).
(C) View of interactions of the hot1-5 position (in CPK coloring) with residues within NDB2 and NBD1 as discussed in the text.
(D) Position of hot1-7 (CPK coloring) relative to the NBD2 ATP binding site. The sensor 2 Arg residue, which is part of the GAR motif, as well as sensor 1 are shown in pink. Residue numbering corresponds to AtHsp101. Figure was prepared with Swiss PDB viewer.