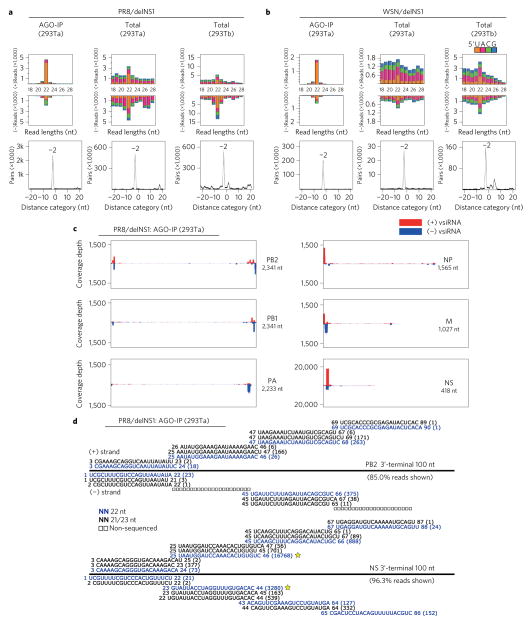
Figure 1. Production of viral siRNAs in mature human somatic cells.
a,b, Size distribution and abundance (per million total mature miRNAs) of total virus-derived small RNAs (vsRNAs) sequenced either directly from two human 293T cell lines (293Ta and 293Tb) 24 h after infection with delNS1 mutants of PR8 and WSN strains (total), or after AGO co-IP from the infected 293Ta cells (AGO-IP). Strong enrichment of total and AGO-bound 22 nt RNAs of both PR8/delNS1 and WSN/delNS1 for pairs (−2 peak) of canonical vsiRNAs with 2 nt 3′ overhangs was detected by computing total pairs of 22 nt vsRNAs with different lengths of base-pairing10. The 5′-terminal nt of vsRNAs is indicated by colour. c, Relative abundance of 21–23 nt vsiRNA hotspots mapped to PR8/delNS1 genomic RNAs, presented from the 3′ end (left) to the 5′ end (right). The genome segments haemagglutinin (HA) and NA, which are targeted by an extremely low density of vsiRNAs, are shown in Supplementary Fig. 1. d, Read sequences along the 3′-terminal 100 nt of PR8/delNS1 mutant genome segments PB2 and NS. Read counts (in brackets), read length, non-sequenced reads, genomic position and percentage of total reads mapped to the region are indicated. The RNAs complementary to the positive (+) or negative (−)-strand vsiRNAs marked by a star were used subsequently as the probes for northern detection of the influenza vsiRNAs.