Fig. 3.
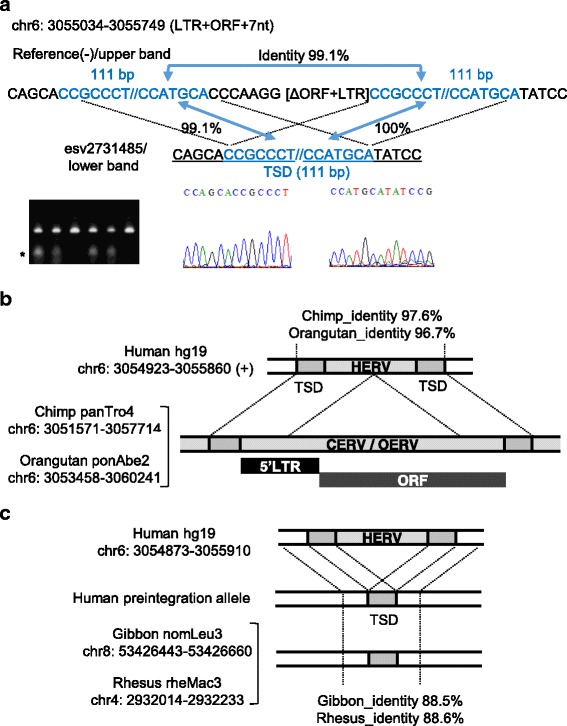
HML-2 structural variation at 6p25.2. a Validation of structural variations at chr6:3,055,034–3,055,749 using Sanger sequencing. Fluorescence data for the underlined sequences are shown below. Blue characters and asterisks show TSD sequences and the presence of preintegration alleles, respectively. b The LTR site at chr6:3,055,034–3,055,749 was an ortholog of ERVs in the chimpanzee (panTro4; chr6: 3,051,682–3,057,596) and orangutan (ponAbe2; chr6: 3,053,569–3,060,123) reference genomes. The regional positions indicated include TSDs. c Putative preintegration alleles corresponding to the LTR site at chr6:3,055,034–3,055,749 in hg19 were assigned in the gibbon (nomLeu3) and rhesus macaque (rheMac3) reference genomes. The regional positions indicated include TSDs and the surrounding regions. Gray regions indicate TSDs. CERV and OERV denote ERV elements in the chimpanzee and orangutan, respectively
